Pone.0034920 1.7
Direct Observation of Strand Passage by DNA-Topoisomerase and Its Limited Processivity
Katsunori Yogo1, Taisaku Ogawa1, Masahito Hayashi2, Yoshie Harada3, Takayuki Nishizaka4,
Kazuhiko Kinosita Jr.1*
1 Department of Physics, Faculty of Science and Engineering, Waseda University, Tokyo, Japan, 2 Yasuda ‘‘On-chip Molecular Cell Phenomics'' Project, Kanagawa Academy
of Science and Technology (KAST), Kawasaki, Japan, 3 Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Kyoto, Japan, 4 Department of Physics,
Gakushuin University, Tokyo, Japan
Type-II DNA topoisomerases resolve DNA entanglements such as supercoils, knots and catenanes by passing one segmentof DNA duplex through a transient enzyme-bridged double-stranded break in another segment. The ATP-dependentpassage reaction has previously been demonstrated at the single-molecule level, showing apparent processivity atsaturating ATP. Here we directly observed the strand passage by human topoisomerase IIa, after winding a pair offluorescently stained DNA molecules with optical tweezers for 30 turns into an X-shaped braid. On average 0.5160.33 mm(1166 turns) of a braid was unlinked in a burst of reactions taking 864 s, the unlinked length being essentially independentof the enzyme concentration between 0.25–37 pM. The time elapsed before the start of processive unlinking decreasedwith the enzyme concentration, being ,100 s at 3.7 pM. These results are consistent with a scenario where the enzymebinds to one DNA for a period of ,10 s, waiting for multiple diffusional encounters with the other DNA to transport itacross the break ,10 times, and then dissociates from the binding site without waiting for the exhaustion of transportableDNA segments.
Citation: Yogo K, Ogawa T, Hayashi M, Harada Y, Nishizaka T, et al. (2012) Direct Observation of Strand Passage by DNA-Topoisomerase and Its LimitedProcessivity. PLoS ONE 7(4): e34920. doi:10.1371/journal.pone.0034920
Editor: Claudine Mayer, Institut Pasteur, France
Received November 19, 2011; Accepted March 7, 2012; Published April 9, 2012
Copyright: ß 2012 Yogo et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permitsunrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: This work was supported by Grants-in-Aid for Specially Promoted Research (grant numbers 16002013, 21000011 to KK) from the Ministry of Education,Culture, Sports, Science and Technology of Japan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of themanuscript.
Competing Interests: The authors have declared that no competing interests exist.
* E-mail:
[email protected]
topoisomerase, the bead suddenly floated upward, indicatingresolution of the entanglements by topoisomerase. These early
Type II DNA topoisomerases let one segment of double-
studies suggested that the enzyme is highly processive. Charvin et
stranded DNA pass through another without leaving a permanent
al. [7], for example, reported that 40- or 80-turn braids they
cut in either of the segments, thus resolving DNA entanglements in
formed were completely unlinked in one burst of reactions in all
cells [1–4]. The multi-subunit enzyme (dimeric in eukaryotes) is
cases examined. Such a high processivity would imply that,
twofold symmetric, and the interface between the two halves
throughout the burst, both of the two DNA molecules can interact
consists of three gates (N-, DNA-, and C-gates) that can open and
with the (single) enzyme molecule simultaneously. Whether the
pass a DNA duplex [5]. In the current view, the enzyme first binds
DNA geometry beneath the bead would allow these simultaneous
one DNA segment, termed the G (gate) segment, at the DNA-gate
encounters was not directly confirmed in the previous studies.
to cleave the double helix while keeping the resultant 59-ends
A particular concern is that a magnetic bead in a magnetic field
bonded to the catalytic tyrosines on each side of the DNA gate.
assumes a certain orientation peculiar to each bead (hence the
When another DNA segment (T or transport segment) comes, the
bead can be rotated by rotating the field). Thus, when two DNA
enzyme transports it sequentially through the three gates in ATP-
molecules of identical lengths are attached to one magnetic bead
dependent reactions, resulting in the passage of the T segment
and the bead is pulled by a magnet, one of the two DNA molecules
across the G segment. Passages of multiple T segments seem to be
must be slack in almost all cases, becoming taut only after a
allowed before the religated G segment eventually leaves the
variable amount of braiding and even then the tension will be
enzyme. How the apparent processivity is regulated, as well as the
different between the two DNAs. Complete unwinding of a loose
precise control of T segment passage without dissociation of the Gsegment from the enzyme or of the dimeric enzyme itself, are yet
braid will be difficult to discern unless one observes the DNAs
to be clarified.
directly. In the case of a single DNA tether, disappearance of
The action of type II topoisomerases has been studied at the
supercoils is directly reflected in the bead movement, but
single-molecule level, with DNA tethering a micron-sized bead to
supercoils are highly dynamic and thus their geometry is not well
a surface [6–9]. Rotating the bead introduced supercoils
(plectonemes) in a single DNA tether, or a braid in a dual DNA
Here we visualized DNA with fluorescence staining, and made a
tether, both resulting in lowering of the bead. In the presence of
braid between a pair of DNA molecules by manipulating each
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Strand Passage by DNA-Topoisomerase
DNA independently. The braid was at the center of an X-shaped
from the trap (Figure 1B1–3). We then turned on fluorescence
DNA pair (Figure 1) with crossing angles of ,90u. Thus the two
excitation, stretched the DNA pair to confirm the absence of extra
DNA molecules could meet only within, or near an end of, the
DNA segments (Figure 1A5, B5), and manipulated the two beads
braid. With this clearly defined geometry, we found that the
with dual-beam optical tweezers to braid the two DNA molecules
processivity of human topoisomerase IIa (hereafter referred to as
tethering the beads (Figure 1A6, B6). We wound one DNA around
topo IIa) is on average ,10 braid turns, limited by a finite burst
the other 30 times, unless stated otherwise. The resultant braid
reaction time of ,10 s after which the enzyme presumably
contained 29.5 turns, because we stopped when the bead came
dissociates from DNA.
back to the original position, but we call this a 30-turn braid forsimplicity throughout this paper. In most cases winding was
counterclockwise as viewed from above. Occasional clockwisetrials did not show an appreciable difference (see figures below),
Direct Observation of Unbraiding Reaction
and thus we ignore the sense of winding in this paper. After
To braid DNA, we made a sparse lawn of l-phage DNA (one
winding, we lowered the two beads to 160.5 mm (bead center)
molecule per 102–103 mm2) by attaching one end of the 16-mm
above the glass surface, and adjusted the bead positions such that
DNA to a glass surface. We then slowly infused topo IIa several
the two DNA molecules would eventually form an X shape with
times to establish a pM concentration of free and active enzyme.
,90u crossings (Figure 1A7, B7). We then displaced the stage until
The final infusion included SYBR Gold, a fluorescent dye for
the DNA pair formed an X shape (Figure 1A9, 1B9) and the
DNA staining, and polystyrene beads (0.92 mm) to be attached to
tension judged from the bead displacement was 0.9560.26 pN
the floating ends of DNA for manipulation (Figure 1A1). We
(mean6s.d. for 85 DNA pairs; ranged 0.44–1.7 pN). The stage
selected in a bright-field image a relatively isolated pair of beads
movement (,10 mm) typically took 10–20 s, the worst case being
that were appropriately separated (5–15 mm) and undergoing
,60 s (no unbraiding reaction followed in this case) and three
tethered diffusion. Each bead was confirmed to be tethered by one
more cases being ,30 s. We take the end of the stage movement
DNA molecule, by holding the bead in an optical trap and moving
the microscope stage in different directions until the bead escaped
Figure 1. Experimental design. (A) Schematic diagrams showing the experimental procedure where the floating ends of the two DNAs weremanipulated with optical tweezers while their root positions were controlled by stage movement. Numbers correspond to those in B–D. (B)Snapshots of bright-field (1–3; sequential frames) and fluorescence images (4–12; averaged over 30 frames = 1 s). Arrows in 1–3 show the direction ofstage movement. Rectangles in 9 show the regions where the DNA images were fitted with a line to estimate the braid length. The scale bar in 12shows 5 mm (57.5 pixels). (C, D) Time courses of the braid length (C) and the DNA tension sensed by the lower-left bead in B (D). Time 0 is the end ofstage movement. After the processive unbraiding at ,60 s, we slightly increased the tension at ,70 s. Gray dots in C were calculated on imagesaveraged over 30 frames, and further averaging over 120 frames (4 s) shown in blue. Red broken lines show the way the burst time was estimated.
Green horizontal bars in C indicate portions shown in Viedo S1. The total tension in D represents (T 2
x +Ty )1/2, and thus noise, converted to positive
values, dominates over the actual tension when the latter is below the noise level. The tension is essentially zero at stages 4 and 12.
doi:10.1371/journal.pone.0034920.g001
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Strand Passage by DNA-Topoisomerase
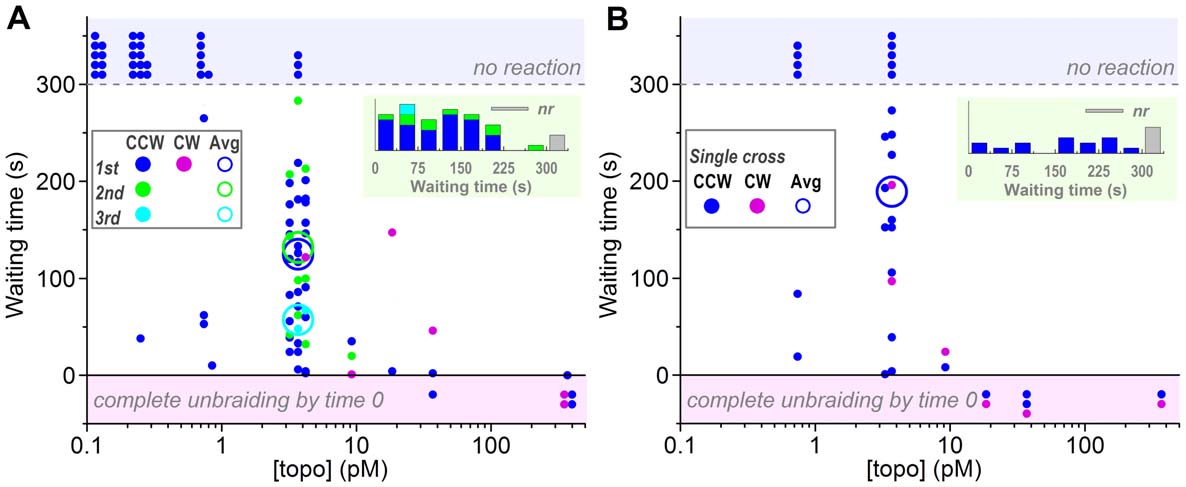
Unbraiding reactions after time 0 were detected as a sudden
times is not possible because times longer than 300 s were not
decrease in the braid length (Figure 1C, 60 s and ,100 s; Video
measured, because short waiting times involve the uncertainties of
S1) and a slight drop in DNA tension (Figure 1D10). In the
10–20 s needed for the stage movement, and because premature
example shown in Figure 1, unbraiding occurred in two bursts,
unbraiding at high [topo IIa]s could not be timed. The trend in
with a decrease of braid length of ,0.7 mm in ,10 s and
Figure 3A, however, suggests that the waiting time is inversely
,1.0 mm in ,8 s. From the relationship between the braid length
proportional to [topo IIa], the relation expected for the scenario
and the number of braid turns obtained in the absence of topo IIa
where binding of one topo IIa molecule initiates each unbraiding
(Figure S3), which depends largely on the crossing angles between
burst. The average waiting time at 3.7 pM topo IIa of ,100 s
the two DNA molecules and to some extent on the tension, we
(excluding no-unbraiding cases and thus being an underestimate)
estimate that the burst lengths above correspond to the releases of
indicates an effective topo IIa binding rate of ,36109 M21s21. A
,13 and ,15 turns, respectively (the sum slightly differs from 30
surface plasmon assay [10] has shown a rate of human
because of imprecision in the calibration and the burst lengths).
After the first burst, we increased the tension slightly by stage
96106 M21s21 at 20uC, while binding of Saccharomyces cerevisiae
movement (Figure 1D10), to confirm that the pause in unbraiding
DNA topoisomerase II to 40 bp (14 nm) DNA assessed through
was not due to the reduction in tension (such a maneuver was
fluorescence anisotropy [11] has given 16109 M21s21 at 30uC.
attempted in nine cases at 3.7 pM topo IIa, all failing to restart
Our value above would be in between for the target DNA size of
unbraiding). The second burst resulted in complete unbraiding, as
1–2 mm (braid length).
seen in the fluorescence image (Figure 1B11), and the tension
When the DNA pair remained intact (not detached from the
dropped to almost 0 (Figure 1D11). The residual small tension due
bead or glass surface and no extraneous DNA on beads) after an
to elongated DNA tether became completely negligible when the
unbraiding assay and subsequent mechanical unbinding, we made
stage was moved back (Figure 1D12). Unless the topo IIa
a single cross (a half-turn braid that we would call one turn in this
concentration, [topo IIa], was high (.,10 pM), complete
paper) with the same DNA pair and waited until topo IIa undid
unbraiding as in Figure 1 was rare: five out of 40 DNA pairs at
the cross or 300 s had passed. Such trials were made up to three
3.7 pM topo IIa and none below. In cases where unbraiding was
times for the same pair. The waiting times for single crosses
incomplete at 300 s, we terminated observation and mechanically
(Figure 3B) were grossly similar to those for 30-turn braids
undid the braid with the optical tweezers, in the absence of
(Figure 3A), indicating that both topo IIa and DNA survived the
fluorescence excitation, to count the number of turns that
rather long (but low intensity) fluorescence excitation. The waiting
remained in the braid. The remaining number was consistent
times for single crosses, however, are longer on average,
with the braid length in most cases, with exceptions where the
particularly when the no reaction cases are taken into account.
mechanical count was obviously smaller (Figure S4). We ascribe
This could be due to the long fluorescence excitation, but a more
the latter to topo IIa-induced unbraiding during mechanical
likely explanation is the smaller target size for topo IIa binding. In
unwinding that took time (the presence of a braid had to be
a DNA cross, the two DNAs slide past each other by thermal
checked after each turn).
motion, and binding of a topo IIa molecule within the thermallyoverlapping zone will be effective. The effective overlap is on the
Size of Unbraiding Bursts
order of 0.2 mm (Text S1), several times smaller than the length of
At 3.7 pM topo IIa, the length of a braid released in one burst
a 30-turn braid.
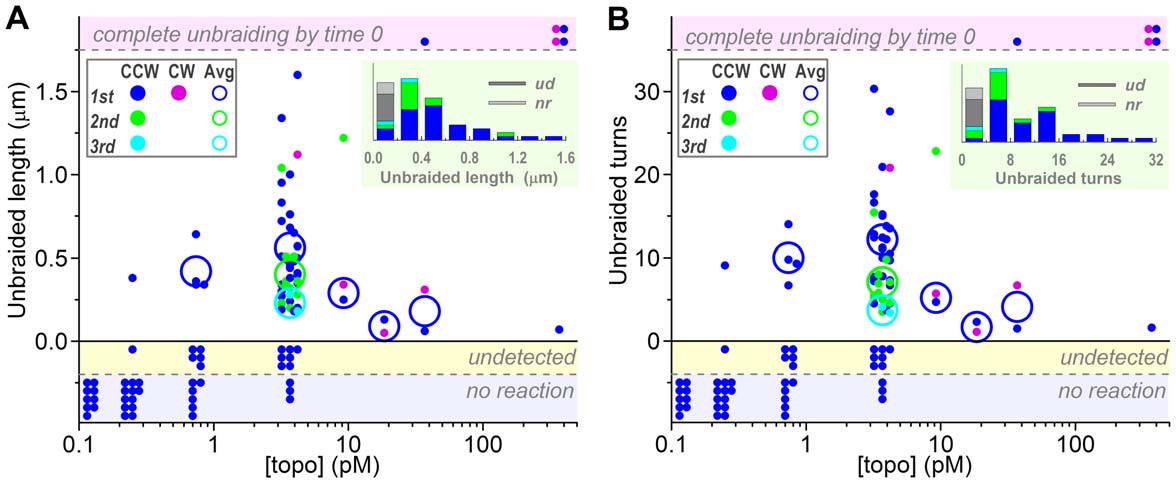
averaged 0.5160.33 mm (s.d. for 43 bursts), corresponding to thenumber of unbraided turns of 10.566.2 (Figure 2). The second
Duration of Unbraiding Bursts
and third reactions in multiple burst cases resulted in progressively
We estimated the duration of each unbraiding burst by drawing
smaller burst sizes (green and cyan in Figure 2): the first bursts
three straight lines, as shown in Figure 1C, to define the beginning
averaged 0.5660.35 mm or 12.266.4 turns (n = 30) whereas the
and end of the burst. We did not find obvious [topo IIa]
second bursts 0.4060.24 mm or 7.063.3 turns (n = 11) and the
dependence, and obtained 864 s (ranged 2–22 s; n = 48, ambig-
thirds 0.2360.07 mm or 3.760.5 turns (n = 2). At lower [topo
uous cases omitted) for the burst duration. In each burst, strand
IIa]s, the unbraiding reactions became rare, but the burst size did
passages took place at a rate of ,1 s21. Burst times of ,10 s or
not change appreciably, suggesting that each burst was catalyzed
less have been reported for relaxation of supercoiled DNA [6].
by one topo IIa molecule. At 3.7 pM topo IIa, the braid length attime 0 was sometimes clearly shorter than that expected for 30
turns (Figure S4), indicating partial unbraiding during the tensionapplication by stage movement or possibly earlier. The premature
In summary, our results show that human topoisomerase IIa
unbraiding was more extensive at higher [topo IIa]s, where, in
unlinks a DNA braid of crossing angles ,90u and under ,1 pN of
most cases, the braids that had remained at time 0 were already
tension in bursts of ,0.5 mm or ,10 turns, each taking ,10 s.
short and disappeared in one short burst. At 370 pM, no braid
One topoisomerase molecule seems to mediate each burst, by
remained by the time ( = 0) the tension was to reach ,1 pN in four
staying on one strand of DNA and allowing multiple passages of
out of five cases (Figure 2, top shaded zone), and winding for 100
the other DNA for a period of ,10 s. These results are largely
turns did not improve this situation. Presumably, the premature
consistent with previous single-molecule unlinking assays [6–9],
unbraiding occurred as rapid succession of multiple bursts rather
except that our [topo IIa] is an order of magnitude lower for
than one burst that encompassed the whole braid.
similar reaction kinetics. We tried to minimize possible loss/damage of topo IIa on surfaces during dilutions and infusions,
Frequency of Unbraiding Bursts
which may partly explain the difference.
In contrast to the burst size which was essentially independent of
All single-molecule assays including the present study have
[topo IIa], the waiting time before an unbraiding burst, whether
shown processive action of topoisomerase II for a duration of the
from the tension setup to the first unbraiding or between two
order of 10 s. A recent study with fluorescence energy transfer [12]
successive unbraiding bursts, decreased greatly as [topo IIa] was
has shown that, once topoisomerase II (Drosophila) binds a DNA
increased (Figure 3A). Precise quantitative analysis of the waiting
segment, it undergoes many MgATP-dependent cycles in which
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Strand Passage by DNA-Topoisomerase
Figure 2. Unbraiding burst sizes. (A) Burst sizes estimated from the braid length in fluorescence images. (B) Burst sizes in terms of braid turns;the braid length was converted to braid turns using the calibration equation described in Figure S3. For both panels, dots show individualobservations, plotted in two or three lines for clarity. Large circles show averages for the first (blue or purple), second (green), and third (cyan) bursts,counterclockwise (CCW) and clockwise (CW) braids not distinguished. The averages are for the data in which unbraiding was confirmed as a changein braid length (dots in the central white zone). Dots in the top shaded zone indicate cases where a braid completely disappeared by the time thetension was set up (time 0). The shaded zone below vertical zero shows cases where a length change was undetected but mechanical unwinding atthe end (300 s) revealed remaining braid turns of less than 30. The bottom shaded zone is for no reaction cases where the braid number remained 30until 300 s as confirmed by mechanical unwinding. Insets at top right show histograms of unbraided length/turns at 3.7 pM topo IIa; ud, undetected;nr, no reaction.
doi:10.1371/journal.pone.0034920.g002
the two ends of the cleaved G-segment DNA are widely separated
KCl+100 mM NaCl) which would favor processivity [13], that
for ,1 s and then come close (and possibly religated) for another
their DNAs were crossed at shallower angles and attached to one
,1 s. Such a behavior, observed in the absence of a T segment to
bead (one of the two DNAs might have been slack, precluding the
be transported, would warrant processivity in that topoisomerase
detection of complete unbraiding), that they worked at a higher
II residing on a G segment is ready to transport through it many
[topoisomerase] of ,30 pM, and that their enzyme was of
other T segments when they come by diffusion. In contrast to the
Drosophila melanogaster. For a single topoisomerase molecule to
report by Charvin et al. [7], however, total unbraiding in one burst
completely undo a braid in an X-shaped DNA pair with 100%
was rare in our experiments. We have no definite explanation,
success, the enzyme would have to somehow move along the DNA
except to point out that they worked at somewhat lower ionic
not randomly but in the direction toward the braid center (see
strength (50 mM KCl+50 mM NaCl compared to our 50 mM
Figure 4A). DNA, however, is basically symmetric, and topoisom-
Figure 3. Waiting times before an unbraiding reaction starts, measured from time 0 (for first bursts) or from the preceding burst(second and third bursts). Observations were terminated at 300 s. (A) Experiments with a 30-turn braid shown in Figure 2. (B) Experiments with asingle-cross braid, attempted after an experiment in A when the DNA pair remained intact. See Figure 2 for symbols. Note that the averages areshown only for [topo IIa] = 3.7 pM, because the cases of no reaction (top shaded zone) or premature unbraiding (bottom shaded zone) introduce toomuch ambiguity. In the averages for the first bursts, the waiting times for the no reaction cases are taken as 300 s, leading to underestimates. Secondand third bursts are also underestimated because of the 300-s limitation in the total observation time. Insets at top right show histograms of waitingtimes at 3.7 pM topo IIa; nr, no reaction.
doi:10.1371/journal.pone.0034920.g003
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Strand Passage by DNA-Topoisomerase
entanglements is physiologically more advantageous than stayinglong on one segment.
Materials and Methods
We purchased recombinant human topoisomerases IIa (NCBI
RefSeq NP_001058.2, unmodified, 340 kDa as homodimer) fromAmersham Pharmacia (USB Product No. 78303Y) and confirmedits stock concentration reported by the supplier of 0.4 mg/mL byBradford assay with BSA as standard. We confirmed in bulk assays(Figure S1) that the enzyme relaxed negatively supercoiledpBR322 plasmid DNA (Nippon Gene) with kinetics similar to orslightly faster than reported [13], that the activity was blocked bythe omission of ATP or by the inhibitor ICRF-193 (Enzo LifeSciences) as described [16], and that the enzyme introduced adouble-stranded break in the plasmid DNA in the presence ofetoposide (TopoGEN) to produce linear DNA [17]. For dilution ofthe enzyme stock, we used non-adsorbing tips and tubes,discarding first dilutions. We labeled ends of unmethylated l-phage DNA (48.5 kb, Promega) with single digoxigenin and single
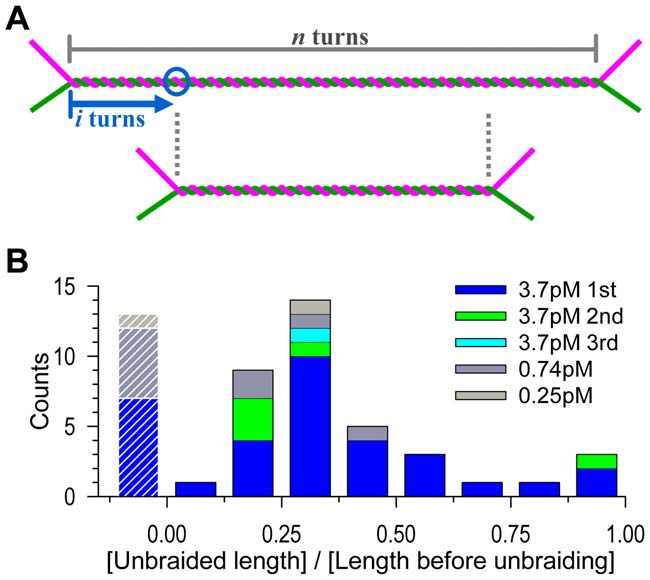
Figure 4. Distribution of unbraiding burst sizes. (A) The expected
biotin through 12-mer primers. Carboxylated polystyrene beads
length of unbraiding. If a topo IIa molecule binds at i-th turn (circle)
(0.92 mm, Bangs) were biotinylated and streptavidin was bound
from an end of a braid of n turns, and if the topo IIa stays and remains
active for a sufficient period, then the braid length will become n – 2i, orthe unbraiding length will be 2i, because the two DNA segmentsforming the braid can freely slide against each other to keep the braid
Observation Chamber
center at the same position. For random binding, i is anywhere between
A ,10 mL flow chamber was made of two cover slips, methanol
0 and n/2, and thus the unbraiding length 2i will distribute equally
cleaned and collodion coated [19]. Infusions (in chamber volumes)
between 0 and n, averaging n/2. Thermal motion of DNA will increase
and incubations were made as follows: 1 volume of base buffer
the unbraiding length by several turns (Text S1). (B) Observed
(10 mM Tris pH 7.9, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2,
distribution. The unbraiding lengths in Figure 2A are normalized by
0.1 mM EDTA); 5 min; 1 vol. 20 mg/mL anti-digoxigenin
the length before unbraiding (corresponding to n in A). Data with aninitial length greater than 1 mm have been selected and analyzed. The
antibody (Roche) in base buffer; 5 min; 3 vol. 1 mg/mL N,N-
leftmost bars with stripes represent cases where unbraiding was
dimethylated casein (Sigma) and 0.2% Tween-20 in base buffer for
undetected in the length assay; the normalized unbraiding lengths for
surface blocking; 5 min; 3 vol. topo buffer (3 mM ATP, 5 mM
these data should be less than 0.25 (Figure S4).
DTT, 0.1 mg/mL dimethylated casein, 0.2% Tween-20 in base
buffer) for washing; 1 vol. 5 pM labeled DNA in topo buffer;15 min; 5 vol. topo buffer containing a desired amount of
erase itself is twofold symmetric [5]. Or, the topoisomerase may
topoisomerase; 1 vol. the same solution containing ,107
catch up a shrinking braid by preferential binding to DNA
streptavidin beads/mL and 800,000-fold dilution of SYBR Gold
crossovers [14], but this would require dissociation before each
(Molecular Probes). Finally the chamber was sealed with grease.
All solutions were prepared with degassed water. Bulk relaxation
Our results here suggest that the processive action of topo IIa is
activity was not affected by SYBR, casein, and Tween-20 (Figure
limited in time. If a topo IIa molecule on a DNA segment can
S1). We also checked decatenation activity with kinetoplast DNA
execute unlimited cycles of strand passages, the reaction would
as substrate (http://www.topogen.com/A_Decatenation.pdf). The
stop when an end of the braid reaches the topo IIa binding site
decatenation activity was unaffected by the presence of SYBR
(Figure 4A). On average, then, 15 turns would be unbraided out of
Gold at 10,000-fold dilution (3467 arbitrary unit, mean6s.d. for
the initial 30 turns, and thermal sliding of the DNAs against each
n = 4) or 0.2% Tween-20 (3162, n = 2) compared to control
other (Text S1) would allow a few more turns to be released. The
(3766, n = 5).
experimental average, in contrast, was 9.6 turns for the 48 firstbursts at [topo IIa] = 3.7 pM (Figure 2B), the undetected cases
Microscopy and Manipulation
being included. It seems that topo IIa does not always wait until a
Fluorescence image of DNA and bright-field image of beads
braid end comes. Figure 4A also shows that the distribution of
were simultaneously observed on an inverted microscope (IX 70,
burst sizes would be uniform if topo IIa stays and remains active
Olympus) equipped with dual-beam optical tweezers (1064 nm)
for an indefinite time. Experimental distribution (Figure 4B),
[20]. Fluorescence was detected with an intensified (VS4-1845,
although the statistics is not good enough to allow the rigorous
Videoscope) CCD camera (R300, Dage-MTI), and beads with
analysis suggested [15], is biased toward short bursts, again
another CCD camera (R300), both at 30 Hz. Two beads tethered
pointing to a limited activity; note that very short unbraiding
to a pair of DNA molecules were manipulated by moving the
bursts were likely unnoticed in our length measurement whereas
trapping beams manually or under computer control with which
some of the long ones may actually have been effected by two topo
one DNA could be wound around the other for 30 turns in 7 s.
IIa molecules. The implication is that, after the average burst
The other ends of the DNAs were manipulated by moving the
period of ,10 s, topo IIa dissociates from DNA (or possibly lapses
motorized sample stage. Stiffness of the optical traps was estimated
into an inactive state), rather than waiting for the exhaustion of
from the (Gaussian) distribution of the positions of a trapped bead
transportable DNA. Perhaps, visiting many different DNA
with a fast-shutter (1 ms) camera (FC300M, Takenaka System) ata reduced laser intensity. Observations were made at 25uC.
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Strand Passage by DNA-Topoisomerase
would fail at Da.45u where the magenta and green rods would
Centroids of bead images were calculated [21] in real time with
overlap partially, although R here represents an entropic radius of
Video-Savant software (IO Industries), which also recorded
DNA [22] which is much larger than the geometrical radius of
digitized images on a hard disk. Length of a DNA braid was
,1 nm and thus the failure would not be a sudden one expected
estimated from the recorded images, after averaging over 30
for solid ropes.
frames (1 s), by fitting each of the four branches of the X-shaped
DNA pair with a line with weights proportional to the fluorescence
Determinants of braid length. Pairs of DNA
intensity (Figure 1B9). Static images (also averaged over 30 frames)
were braided and the braid lengths estimated from fluorescence
were also fitted with lines by eye, after magnification, giving
images as described in the main text, except that topoisomerase
similar results. Calibration in the absence of topo IIa (Figure S3)
was not included in the medium. (A) Fluorescence images of a
indicates an average precision of 60.12 mm (61.4 pixel).
DNA braid of n = 30, showing dependence of the braid length onbraid angles. (B) Proportionality between braid length and braid
Supporting Information
turns (n – 1/2) for h around 90u (range 75u–95u). (C) Angle
Bulk supercoil relaxation activity of topo IIa.
dependence of the braid length for n = 30. The lower horizontal
The enzyme at the indicated concentration was incubated with
axis is chosen to test the geometrical model in Figure S2. Open
5 nM pBR322 plasmid (negatively supercoiled) and 1 mM ATP
circles, tension between 0.7 and 1.4 pN; black circles, below
(unless indicated otherwise) for the indicated time at 37uC in
0.7 pN; gray circles, above 1.4 pN. Dashed line shows regression
20 mL of buffer B for bulk assay or buffer M for microscopy. Buffer
passing through the origin (geometrical model) for open circles,
B was the base buffer described in the main text (10 mM Tris
with l = 1.37tan(90u - h/2) where l is the braid length in mm. A
pH 7.9, 100 mM NaCl, 50 mM KCl, 5 mM MgCl
better fit is obtained if we allow the line to deviate from the origin
EDTA) containing, in addition, 0.1 mg/mL BSA. For buffer M,
(solid line), with l = 0.51+0.96tan(90u - h/2). Deviation from the
the base buffer was supplemented with 5 mM DTT, 0.1 mg/mL
geometrical model (Figure S2) is expected for h.90u, and the
dimethylated casein, 0.2% Tween-20, and 800,000-fold dilution of
observed braid pitch of ,50 nm (l/n), close to the persistence
SYBR Gold. The reaction was started by the addition of 1 mL of
length of DNA, also suggests that bending the DNA may cost
the enzyme diluted in buffer B containing 0.5 mM DTT, and
additional energy. (D) Tension dependence of the braid length
terminated by the addition of 2 mL of 5% SDS and 100 mM
(n = 30) for h around 90u (range 79u–97u). (E) Braid lengths in D
EDTA. Samples were mixed with 2 mL of gel loading buffer (50%
converted to those at h = 90u by assuming the angle dependence
glycerol, 0.9% SDS, 0.05% Bromophenol Blue), heated at 70uC
shown in the solid line in C. Solid curve shows fit assuming that
for 2 min, subjected to electrophoresis in a 1% agarose gel in
the normalized braid length depends on the tension F as F23/4 [7]:
90 mM Tris-borate pH 8.4 and 2 mM EDTA, and stained with
l = 0.90+0.63F23/4 where F is in pN. In the main text and in
10,000-fold dilution of SYBR Gold. (A) Time courses of
Figure S4 below, we estimate the braid turns n from the observed
relaxation. (2)SC, negatively supercoiled plasmid; relaxed, relaxed
braid length l and tension F assuming this tension dependence and
circular plasmid. (B) Effects of topoisomerase-specific drugs.
the solid line in C: l = [(n – 1/2)/(29.5?1.47)][0.51+0.96tan(90u -
ICRF-193, a bis(2,6-didioxopiperazine) derivative, is a potent
n = 1/2+72l/{[0.53+tan(90u
inhibitor of type II topoisomerase with 50% inhibition at ,2 mM
2)](1.43+F23/4)}. The root-mean-square deviation of the 58
[16]. Etoposide inhibits religation of DNA, producing double-
measured braid lengths from this phenomenological equation
strand breaks irrespective of the presence of ATP [17]. Assays
(four data with tan(90u - h/2),0.5 excluded) is 0.12 mm, which is a
were made as in A for the reaction period of 30 min. Samples with
measure of the reliability of the length estimates.
etoposide were incubated, prior to electrophoresis, with 200 mg/
mL proteinase K at 45uC for 30 min to digest topo IIa. Linear
Summary of individual unbraiding reactions.
DNA marker was generated by digesting pBR322 by EcoRI. The
Results from each experiment are shown in paired bars, the left
yield of linearized DNA at 200 mM etoposide is below 10% at the
bar showing the braid turns counted by mechanical winding/
enzyme/pBR322 molar ratio of five under similar conditions [17].
unwinding and the right bar showing the observed braid lengths
which have been converted to the braid turns by the last equation
A geometrical model for a DNA braid. The
in Figure S3. All the left bars are given the same height of 30 turns,
model has been described by Charvin et al. [7] for the case of a
the count in the initial mechanical winding. Note that the count at
symmetric braid (Da = 0). Here, the angles between the DNA and
time 0 may have been smaller, due to premature unbraiding
braid axis are a+Da (magenta) and a-Da (green), which are
particularly at high [topo IIa]s. The magenta portions show the
assumed to be maintained through the braid. In the braid, the
braid turns that remained at 300 s, estimated by mechanical
distances between the DNA axis and braid axis are taken as R+DR
unbraiding. Some of these may be underestimated, due to
(magenta) and R-DR (green). The pitch P is then given as
unbraiding by topo IIa during the process of mechanical
P = 2p(R+DR)/tan(a+Da) for magenta DNA and P = 2p(R – DR)/
unbraiding. The height of the bars on the right corresponds to
tan(a – Da) for green DNA. The two pitches must be the same in a
the braid length at time 0. The height deviates from 30, because
braid, leading to P = 2pRcota(1 - Da2/cosa2) where terms higher
the determination of the braid length and the conversion to braid
than Da2 have been neglected. For a braid of (n – 1/2) turns (made
turns both involve errors, and because premature unbraiding took
by winding one DNA around the other for n turns as described in
place at high [topo IIa]s. For the same reasons, the magenta
the main text), its length l is given by l = (n – 1/2)P. The effect of
portion in the left bar and the red portion in the right bar do not
the asymmetry (Da) is of the second order and is practically
match precisely. ‘‘ud'' indicates the cases where the mechanical
negligible, and thus l,2p(n – 1/2)Rcota = 2p(n – 1/2)Rtan(p/2 -
count suggested unbraiding but a decrease in the braid length
h/2), where h = 2a is the angle between the two DNAs. This final
could not be detected reliably. Note that failure in detecting a
result is the same as that by Charvin et al. [7], and the only merit
length change can be inferred only when the initial length was
of the calculation above is to show that asymmetry does not matter
apparently preserved; when unbraiding is detected both in the
as long as Da,,1 (radian). Note that the geometrical model
mechanical counts and length changes, there is always a possibility
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Strand Passage by DNA-Topoisomerase
that an additional short unbraiding event(s) has taken place
frames, and the intensity adjusted to highlight DNA (bead images
without being detected. CW, clockwise braid.
are saturated).
Unbraiding of a 30(29.5)-turn DNA braid in
DNA fluctuations in a braid.
two bursts. The movie consists of three portions (initial setup,
first unbraiding burst, second burst resulting in completeunbraiding) as indicated by green horizontal bars in Figure 1C
in the main text, with 1-s blackouts in between. The time displayed
We thank members of Kinosita Lab for technical assistance and discussion,
on the lower right matches that in Figure 1C. The white bar that
and S. Takahashi, K. Sakamaki, M. Fukatsu, and H. Umezawa for
appears at time 0 indicates 1.8 mm, the initial braid length. Note
encouragement and lab management.
that the stage, and thus the DNA roots, were slightly moved to theright between ,70 and ,75 s to increase the tension (Figure 1D).
Author Contributions
The movie taken at 30 frames/s has been running-averaged for 30
Conceived and designed the experiments: KY. Performed the experiments:KY TO MH YH. Analyzed the data: KY TO. Wrote the paper: KY YHTN KK.
Liu LF, Liu CC, Alberts BM (1979) T4 DNA topoisomerase: a new ATP-
13. McClendon AK, Rodriguez AC, Osheroff N (2005) Human topoisomerase IIa
dependent enzyme essential for initiation of T4 bacteriophage DNA replication.
rapidly relaxes positively supercoiled DNA. Implications for enzyme action
Nature 281: 456–461.
ahead of replication forks. J Biol Chem 280: 39337–39345.
Brown PO, Cozzarelli NR (1979) A sign inversion mechanism for enzymatic
14. Zechiedrich EL, Osheroff N (1990) Eukaryotic topoisomerases recognize nucleic
supercoiling of DNA. Science 206: 1081–1083.
acid topology by preferentially interacting with DNA crossovers. EMBO J 9:
Mizuuchi K, Fisher LM, O'Dea MH, Gellert M (1980) DNA gyrase action
involves the introduction of transient double-strand breaks into DNA. Proc Natl
15. Koster DA, Wiggins CH, Dekker NH (2006) Multiple events on single
Acad Sci U S A 77: 1847–1851.
molecules: Unbiased estimation in single-molecule biophysics. Proc Natl Acad
Wang JC (1998) Moving one DNA double helix through another by a type II
Sci U S A 103: 1750–1755.
DNA topoisomerase: the story of a simple molecular machine. Q Rev Biophys
16. Tanabe K, Ikegami Y, Ishida R, Andoh T (1991) Inhibition of topoisomerase II
31: 107–144.
by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res 51:
Schoeffler AJ, Berger JM (2008) DNA topoisomerases: harnessing and
constraining energy to govern chromosome topology. Q Rev Biophys 41:
17. Bromberg KD, Burgin AB, Osheroff N (2003) A two-drug model for etoposide
action against human topoisomerase IIa. J Biol Chem 278: 7406–7412.
Strick TR, Croquette V, Bensimon D (2000) Single-molecule analysis of DNA
18. Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, et al. (2001) Direct
uncoiling by a type II topoisomerase. Nature 404: 901–904.
observation of DNA rotation during transcription by Escherichia coli RNA
Charvin G, Bensimon D, Croquette V (2003) Single-molecule study of DNA
polymerase. Nature 409: 113–115.
unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc Natl AcadSci U S A 100: 9820–9825.
19. Hayashi M, Harada Y (2007) Direct observation of the reversible unwinding of a
Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, et al. (2003)
single DNA molecule caused by the intercalation of ethidium bromide. Nucleic
Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type
Acids Res 35: e125.
II topoisomerases. Proc Natl Acad Sci U S A 100: 8654–8659.
20. Arai Y, Yasuda R, Akashi K, Harada Y, Miyata H, et al. (1999) Tying a
No¨llmann M, Stone MD, Bryant Z, Gore J, Crisona NJ, et al. (2007) Multiple
molecular knot with optical tweezers. Nature 399: 446–448.
modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat
21. Yasuda R, Noji H, Yoshida M, Kinosita K, Jr., Itoh H (2001) Resolution of
Struct Mol Biol 14: 264–271.
distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase.
10. Renodon-Cornie re A, Jensen LH, Nitiss JL, Jensen PB, Sehested M (2002)
Nature 410: 898–904.
Interaction of human DNA topoisomerase IIa with DNA: Quantification by
22. Marko JF (1997) Supercoiled and braided DNA under tension. Phys Rev E 55:
surface plasmon resonance. Biochemistry 41: 13395–13402.
11. Mueller-Planitz F, Herschlag D (2008) Coupling between ATP binding and
23. Bustamante C, Marko JF, Siggia ED, Smith SB (1994) Entropic elasticity of l-
DNA cleavage by DNA Topoisomerase II: A unifying kinetic and structural
phage DNA. Science 265: 1599–1600.
mechanism. J Biol Chem 283: 17463–17476.
24. Strick TR, Allemand J-F, Bensimon D, Croquette V (1998) Behavior of
12. Smiley RD, Collins TRL, Hammes GG, Hsieh TS (2007) Single-molecule
supercolied DNA. Biophys J 74: 2016–2028.
measurements of the opening and closing of the DNA gate by eukaryotictopoisomerase II. Proc Natl Acad Sci U S A 104: 4840–4845.
PLoS ONE www.plosone.org
April 2012 Volume 7 Issue 4 e34920
Source: http://www.k2.phys.waseda.ac.jp/PDF/2012PlosOne_Yogo_TopoII.pdf
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Agents that kill bacteria are said to be A) bacteriostatic. B) inhibitory. C) bacteriocidal. D) all of the above. 2) Compared with decontamination, disinfection is
Blei, Lithium oder Natrium: Woraus man die besten Akkus baut, hängt nicht nur von der Chemie ab Das Einmachglas für Elektronen gibt es noch nicht, doch manche Akkus sind schon ziemlich nah dran PHOTON Oktober 2010 Bislang wird überschüssiger Strom vor allem in Pumpspeicherkraft- Elektrischer Strom wird sich nie


