Amgs.or.kr
APEC Youth Scientist Journal Vol.7 / No.2
ACQUIRING OF BACTERIAL ABR STRENGTH BY VARIOUS
CONCENTRATIONS OF ANTIBIOTICS AND SPREADING
PATHWAY OF ABR TRAIT
1Suffield Academy, 185 North Main Street, Suffield, CT 06078 USA
This research was conducted to examine the extent bacteria develop antibiotic
resistance (ABR) from contemporary and old bacteria, and understand its underlying
mechanisms of the process. Common bacteria and B. cereus, S. aureus, E. coli, and S.
gallinarum were cultured with various concentrations of antibiotics and underwent agar
diffusion tests to determine resistance. Then ABR and non-ABR bacteria were cultured in a
single solution and ABR was tested. Plasmid DNA from ABR bacteria were implanted into
non-ABR bacteria and resistance of possibly transformed bacteria was tested. Finally,
plasmid DNA were extracted from ABR bacteria and inserted to non-ABR bacteria of the
same kind through heat shock transformation and resistance determined. Results showed
that common bacteria and B. cereus, S. aureus, E. coli, and S. gallinarum have grown ABR to
old antibiotics and are starting to develop it on newer antibiotics. Non-ABR bacteria
develop stronger ABR under lower concentration of antibiotics through proximity with ABR
bacteria. Also, bacteria transformed with ABR plasmid DNA exhibited an ABR trait. It
can be concluded many bacteria have adjusted to antibiotics, and they have grown this ABR
through exposure to low concentration of antibiotics, proximity with ABR bacteria, and
plasmid DNA transformation from ABR bacteria.
* Corresponded to: Sang On PARK (
[email protected])
APEC Youth Scientist Journal Vol.7 / No.2
1. INTRODUCTION
Antibiotics or antimicrobial drugs are used to cure bacterial causing illnesses. Bacteria
are single celled organisms, and certain kinds are believed to cause illnesses. Although
antibiotics can decimate bacteria, there is a consequence. When bacteria are exposed to
antibiotics, initially, the sensitive majority will be killed; however, there will be surviving
minority, which had different trait to dead ones. Like Darwin's survival of the fittest theory,
these fit bacteria prevail, and the population will be less susceptible to the next exposure.
(Bergman, 2015) Swedish research team further found that heavy usage of antibiotics
increases the mutation frequency in bacteria. (Gustafsson et al., 2015) This antibiotics
resistance nuisance is becoming more serious as antibiotics resistance was found even for
ceftaroline, which was introduced in 2010. (Timeline of Antibiotic Resistance, 2015) Bacteria
are acquiring stronger resistance to antibiotics as old introduction and long exposure is
decreasing antibiotics effectiveness. (Harjvan et al., 2015) Contemporary bacteria have
higher resistance than those from the time before antibiotics were introduced. (Houndt &
Ochman, 2015) There are several classes of antibiotics: penicillin, cephalosporin, macrolides,
fluoroquinolones, sulfonamides, tetracycline, and aminoglycosides. Symptoms are often
bacteria specific, so doctors prescribe appropriate antibiotics accordingly. (Stephens, 2015)
There are two classes of bacteria: gram-positive and gram-negative. When purple dye is
dropped, gram-positive acquire dye's color, but gram-negative are colored red or pink. (A
Brief Overview of Classes of Antibiotics, 2015) Like bacteria there are two big classes of
antibiotics. One is bacteriostatic agent, which inhibits the growth and reproduction, and
another is bactericidal agent, which destroys bacteria. Penicillin, cephalosporin,
fluoroquinolones, and aminoglycosides are considered bacterial agents; macrolides,
sulfonamides, and tetracycline are considered bacteriostatic agents. Science has discovered
means in which bacteria pass down its antibiotics resistance. One way is the inheritance of
plasmid DNA, which will enter other bacteria and make changes accordingly. A University
of Granda made a hypothesis that with indiscriminate antibiotics use, antibiotics put more
stress on bacteria, which will then move more actively and accept more DNA because
bacteria usually do not search for new DNA. (New Hypothesis: Why Bacteria are becoming
Increasingly more Resistant to Antibiotics, 2015) Report on 2007 Science magazine says that
a research team has found stealth gene like Sfh protein. Sfh protein is believed to transmit
plasmid DNA without changing bacteria's fitness much because bacteria had serious change
in fitness when plasmid DNA was transmitted without Sfh protein. (Doyle et al, 2015)
APEC Youth Scientist Journal Vol.7 / No.2
Howard Hughes medical Institute discovered that antibiotic resistant gene could be
transmitted by cell-cell contact and integrating into a chromosome. (Beaber et al., 2015)
Finally, research team from Uppsala University discovered that plasmid DNA that contains
antibiotics resistance can be strengthened by exposure to very low level of antibiotics and
heavy metal. (Gullberg et al., 2015)
Based on these researches five experiments were conducted. First two were to test
ABR trend in common bacteria and stored specific bacteria. Common bacteria from soil were
done agar diffusion test with antibiotics, and stored specific bacteria were done the same agar
diffusion test with same antibiotics. Certain bacteria and antibiotics were selected for further
experiments to figure out mechanisms for ABR spread, for bacteria with strong ABR to
certain antibiotics would be inappropriate for the purpose. Next three experiments were
conducted to propose three possible mechanisms for ABR development. The first one was
exposing bacteria to different concentrations of antibiotics and doing agar diffusion test with
0.001x cef. to find how concentration of antibiotics could play role in ABR development. The
second was cultivating ABR bacteria and non-ABR bacteria in same nutrient broth to find
possible directional shift in both or one regarding ABR development. The last was extracting
plasmid DNA from ABR bacteria and implanting it on non-ABR bacteria to ensure whether
transformation by plasmid DNA could help the ABR development.
2. MATERIALS AND METHODS
Experiment 1: Status of ABR to various antibiotics on different types of soil bacteria
Bacteria samples were made by 0.1 g of three different soil samples, labeled 1 through
3, from different flowerpot mixed with 0.9 mL distilled water in respective 1.5 mL micro
tube. Components were then mixed by vortex mixer and waited for soil to suspend. Mixture
was streaked with 10 micro liter loop on NA. The solution was streaked on the half of the
NA, and dish was turned 90 degrees. The solution was again streaked half of NA from the
perspective, but only upper half was overlapping with previous streaking. Streaking
continued from the second step until NA was completely covered with solution. NA plates
were put in incubator with 37 degree Celsius for 24 hours. Cultivated bacteria were done agar
diffusion test by poking 7 circles in the middle of NA with micropipette white tip, picked up
by tong heated by alcohol lamp. Dropping 10 microliters of prepared antibiotics on each
circle and incubating ends the test. Cefazedone and kanamycin were diluted to 0.1x. Using
APEC Youth Scientist Journal Vol.7 / No.2
micropipettes, 0.9 mL of distilled water was put in a tube and 0.1 mL of 1x antibiotics were
Experiment 2: Acquiring of ABR on bacteria exposed to various antibiotics
8 petri dishes were prepared for each bacterium:
Escherichia coli,
Salmonella
gallinarum,
Bacillus cereus, and
Staphylococcus aureus. 10 micro liters of each bacterial
solution were dropped on all 8 petri dishes and were spread 30 times with a spreader. A circle
was poked on each petri dish and all different eight antibiotics were dropped 10 micro liters
on different bacteria petri dish respectively.
Experiment 3: Different degree of bacterial ABR strength by various concentrations of
antibiotics
Three selected bacteria that are thought to not have resistance to two chosen
antibiotics were done agar diffusion test with different concentrations for cefazedone and
lincomycin: 0.1x, 0.01x, 0.001, and 0.0001x. Mixing 100 microliters of 1x, 0.1x, 0.01, and
0.001x with 900 microliters of nutrient broth in a tube. Bacteria were exposed to the
antibiotics by dropping 10 microliters of bacterial solution into a tube for every bacterium.
After 24 hours of incubation agar diffusion test were done for each bacterium with 5
microliters of 0.001x cefazedone.
Experiment 4: Antagonistic effects between ABR acquired bacteria and normal bacteria
in co-culture
ABR bacteria and non-ABR bacteria were mixed in NB. ABR bacteria were scratched
from experiment 3 samples, specifically ones close to the clear zone. For non-ABR bacteria
stored bacterial solution was used. The NB solution was incubated for 24 hours. There were
three solutions:
E. coli and
S. gallinarum solutions with lincomycin resistant bacteria mixed
and cefazedone resistant
S. gallinarum mixed with stored S. gallinarum. Then, the solution
was done agar diffusion test with 0.001x cefazedone and incubated for 24 hours.
Experiment 5: Possibility of ABR spread by plasmid DNA transfection
Before mixing ABR bacteria from experiment 3 and bacterial solution without ABR
volume for each solution had to be calculated so that the absorbance is 0.1 compared to
nutrient broth. After mixing appropriately the solution was incubated for 24 hours. Then, 20
or 30, depending on clarity, microliters of solution were dropped and spread on NA. NA was
APEC Youth Scientist Journal Vol.7 / No.2
poked with a circle in the middle and 5 microliters of 0.001x cefazedone was dropped. NA
are incubated for 24 hours again.
-
Plasmid DNA isolation; Both no ABR bacterial solution and ABR bacterial
solutions were centrifuged at 13000 rpm for a minute. The remaining liquid is
disposed. 250 microliters of buffer 1 was added to remainders and mixed by vortex
machine. 250 microliters of buffer 2 was added and mixed by inverting the micro
tube 4 times. 350 microliters of buffer 3 were added and mixed by inverting 4 times.
The tube is centrifuged at 13000 rpm for 19 minutes. Cleared lysate was transferred
to DNA binding column tube and centrifuged at 13000 rpm for a minute. Liquid was
disposed and 700 microliters of buffer 4 was added to DNA binding column tube.
The tube was centrifuged at 13000 rpm for a min. Liquid was again disposed and the
filter was dried of residual ethanol by centrifugation at 13000 rpm for a minute. The
filter as moved to 1.5 mL micro centrifuge tube, and 100 microliters of buffer 5 or
distilled water were added to DNA binding filter column and centrifuged at 13000
rpm for a minute for elution. DNA is ready.
-
Heat shock transformation; To combine DNA, 2 mL of bacteria were put in ice for
10 minutes first. Then, bacteria were centrifuged at 4500 rpm for 10 minutes. Liquid
formed will be disposed, and 1 mL of 0.1M CaCl2, made by mixing 0.50 g of CaCl2
and 45 mL of distilled water, was dropped in and mixed by inverting. The bacteria
are then put in ice for 15 minutes. Then, bacteria are centrifuged at 3500 rpm for 10
minutes. Liquid is disposed again, and 200 mL of previous CaCl2 solution was
added. Again, bacteria are put in ice for 30 minutes, 100 microliters of plasmid DNA
are dropped in. The tube is put in 42 degree Celsius for 2 minutes and at 0 degree
Celsius for 2 minutes. 9 mL of nutrient broth was put in, and bacteria were incubated.
After planting plasmid DNA of ABR bacteria to no ABR bacteria and incubating for
24 hours the solution was dropped 20 to 30 microliters, depending on the clarity, on NA and
did diffusion test with 0.001x cefazedone.
APEC Youth Scientist Journal Vol.7 / No.2
3. RESULTS
Experiment 1: Status of ABR to various antibiotics on different types of soil bacteria
Different bacteria from each soil sample were done agar diffusion test to observe how
much ABR did common bacteria grown. Classifying of bacteria was based on the size and
color of soil bacterial colonies within the same soil sample. While most or all bacteria had
resistance to relatively old antibiotics like ampicillin, methicillin, chloramphenicol, and
penicillin, which means they had clear zone diameter of only initial hole for agar diffusion
test, only 3 had resistance to cefazedone. All others had increase in clear zone diameter,
ranging from 1.3 cm in soil sample 2 bacteria d1 to 4.8 cm increase in bacteria e from soil
sample 3. Additionally, only 5 had resistance for kanamycin, and only 1 had resistance for
lincomycin. The most resistant without ABR was soil sample 1 bacteria b2 with diameter of
0.8 cm, and the least resistant was soil sample 1 bacteria c2 with diameter of 1.8 cm. The
least resistant to lincomycin was soil sample 1 bacteria a with 4.7 cm diameter and most
resistant was soil sample 2 bacteria c2 with diameter of 0.8 cm. Antibiotics with low
resistance were introduced later than those with high resistance. Penicillin and
Chloramphenicol were introduced in between 1940 and 1950. Methicillin and Ampicillin
were introduced around 1960. Cefazedone, kanamycin, and lincomycin were introduced after
2000. The conclusion can be drawn that common bacteria like those in the soil have
developed resistance to old antibiotics over time, but did not form resistance to newer
antibiotics. They are growing ABR, however, as evident in some bacteria having strong or
ABR to modern antibiotics.
APEC Youth Scientist Journal Vol.7 / No.2
Table 1: Diameter of clear zone to various antibiotics on soil bacteria
Experiment 2: Acquiring of ABR on bacteria exposed to various antibiotics
Four bacteria,
E. coli,
S. gallinarum,
B. cereus, and
S. aureus, were done agar
diffusion test with seven different antibiotics to figure which bacteria has ABR to which
antibiotics thus selecting appropriate bacteria and antibiotics for experiment 3. Resistance
was measured in diameter of the clear zone. While
S. gallinarum did not have resistance for
all antibiotics,
E. coli,
B. cereus, and
S. aureus all had resistance for all antibiotics except
cefazedon, kanamycin, and lincomycin. All four bacteria had resistance to cefazedone.
E. coli
had 5 cm clear zone.
S. gallinarum had 3.4 cm and
B. cereus had 5.4 cm.
S. aureus had 8.5
cm clear zone. Bacteria for next experiment were selected; they were all bacteria except
S.
aureus because it had the lowest resistance to antibiotics that all bacteria did not have
resistance to. This is shown in kanamycin and lincomycin as well because
S. aureus had 1.8
cm clear zone and 5.4 clear zone respectively.
E. coli had 1.1 cm and 3 cm;
S. gallinarum had
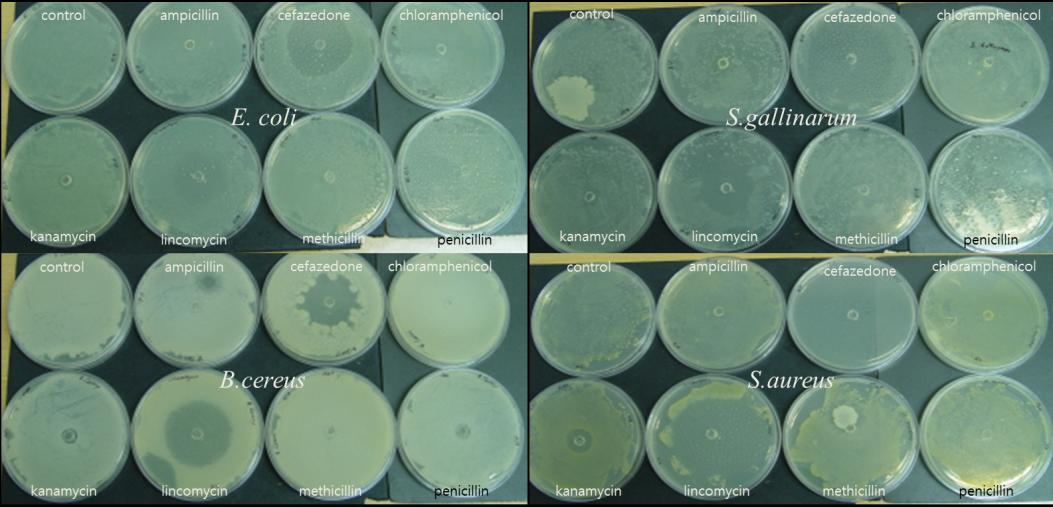
APEC Youth Scientist Journal Vol.7 / No.2
1.1 cm and 4.4 cm; B cereus had 1 cm and 3.7 cm respectively. Antibiotics were selected
cefazedone and lincomycin because even though bacteria did not form complete resistance
against kanamycin, they had the smallest clear zone with kanamycin, suggesting that they
formed quite robust but not complete resistance. Conclusion can be drawn that E. coli, B.
cereus, and S. aureus are appropriate bacteria, and cefazedone and lincomycin, which
bacteria not yet have resistance but have smaller clear zone, are appropriate antibiotics for
next experiment.
Table 2: diameter of bacterial clear zone according to various antibiotics
Figure 1: Clear zones of four bacteria with different antibiotics
Experiment 3: Different degree of bacterial ABR strength by various concentrations of
antibiotics
To figure out whether bacteria exposed to cefazedone acquires antibiotics resistance
to cefazedone, three kinds of bacteria, E. coli, S. gallinarum, and B. cereus, were cultivated
with various concentrations of antibiotics and did agar diffusion test to cefazedone S.
gallinarum had identical size from 0.1x cefazedone to 0.001x cefazedone of 8.5 cm, but on
0.0001x cefazedone clear zone diameter of 8.5 cm, nearly a diameter petri dish, dropped to 5
cm. B. cereus, like S. gallinarum, had similar size up to 0.001x cefazedone of 7.1 cm, 6.4 cm,
APEC Youth Scientist Journal Vol.7 / No.2
and 7.3 cm, but at 0.0001x cefazedone, its clear zone decreased from 7.3 cm to 5.8 cm. E.
coli had different pattern. As the concentration got dimmer the size of clear zone dwindled.
From 0.1x cefazedone to 0.01x cefazedone, diameter dropped from 8.5 cm to 6.6 cm.
Transition from 0.01x to 0.001x did not affect the diameter, but diameter dropped from 6.6
cm to 4 cm as concentration dropped from 0.001x cefazedone to 0.0001x cefazedone. All
three kinds of bacteria exposed to cefazedone had decrease in clear zone size at the lowest
concentration of cefazedone. With the exception of S. gallinarum two bacteria showed drop
in diameter at first change in concentration of cefazedone. On second change of concentration
E. coli and S. gallinarum maintained same diameter while B. cereus had increased diameter
from 6.4 cm to 7.3 cm. Therefore, result can be concluded that low concentration of
antibiotics can incur stronger ABR.
Figure 2: Diameter of clear zone for three bacteria with different concentration of cef.
To find whether bacteria exposed to lincomycin will have ABR to different antibiotics
like cefazedone, E. coli, S. gallinarum, and B. cereus, previously cultivated with various
concentrations of lincomycin, were on agar diffusion test to cefazedone. S. gallinarum's size
of clear zone dropped from 8.5 cm to 2.6 cm as the concentration changed from 0.1x
lincomycin to 0.01x lincomycin, and the size of clear zone maintained similar diameter for
0.01x lincomycin, 0.001x lincomycin, and 0.0001x lincomycin. of 2.6 cm, 2.2 cm, and 2.4
cm. B cereus had similar pattern of dropping from 6.6 cm to 4.5 cm from 0.1x lincomycin to
0.01x lincomycin and maintained similar clear zone size for 0.01x lincomycin, 0.001x
lincomycin, and 0.0001x lincomycin of 4.5 cm, 4.9 cm, and 4.9 cm. E. coli maintained
similar clear zone size for 0.1x lincomycin, 0.01x lincomycin, and 0.001x lincomycin of 3.6
cm, 3.2 cm, and 3.1 cm, but as the concentration shifted from 0.001x lincomycin to 0.0001x
lincomycin, the diameter of clear zone dropped from 3.1 cm to 1.5 cm. Therefore, the ABR
APEC Youth Scientist Journal Vol.7 / No.2
developed at lower concentration of antibiotics is stronger, and despite the discrepancy in
kinds of antibiotics, ABR can be acquired to newly contacted antibiotics.
Figure 3: Diameter of clear zone with different concentration of lin.
Experiment 4: Antagonistic effects between ABR acquired bacteria and normal bacteria
in co-culture
S. gallinarum with ABR to licomycin from experiment 3 was mixed with non ABR S.
gallinarum and incubated to observe effect of ABR bacteria have on non ABR bacteria. The
diameter of ABR bacteria and non ABR bacteria decreased from 2.4 to 1 and from 5.4 to 3
respectively. Decrease in clear zone means that both grew ABR to cefazedone, and the
difference in clear zone decreasing from 3 to 2 concludes that the antagonistic effect of two
bacteria was applied. Additionally, control S. gallinarum has dominance over ABR S.
gallinarum.
Same thing was done except with cefazedone ABR bacteria for this one. Diameters of
clear zone decreased from 5 to 0.7 and 5.4 to 3.8 for ABR bacteria and control bacteria
respectively. The general increase in ABR was observed, but increase in difference of clear
zone from 0.4 to 3.1 shows that ABR bacteria are dominant over control.
Same thing was done except with control E. coli and licomycin ABR E. coli.
Diameters of clear zone decreased from 1.5 to 1.3 and 5 to 3.2 for ABR bacteria and control
respectively. The difference of clear zone decreased from 3.5 to 1.9, so conclusion can be
drawn that control has dominance over ABR bacteria in antagonistic effect.
APEC Youth Scientist Journal Vol.7 / No.2
Figure 4: Diameters of lin. ABR and control bacteria clear zone before and after co-
Figure 5: Diameters of cef. ABR transformed and control bacteria clear zone
before and after co-cultivation
Figure 6: Diameters of lin. ABR transformed and control bacteria clear zone
before and after co-cultivation
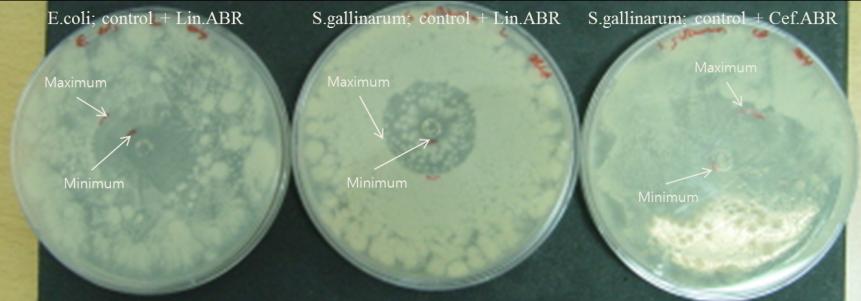
APEC Youth Scientist Journal Vol.7 / No.2
Figure 7: Cultivation of control and ABR transformed bacteria
Experiment 5: Possibility of ABR spread by plasmid DNA transfection
To confirm that ABR can be spread with plasmid DNA, heat shock transformation
was used to implant plasmid DNA from ABR E. coli and S. gallinarum to non ABR E. coli
and S. gallinarum respectively. Transformed bacteria were done agar diffusion test. S.
gallinarum showed resistance to both lincomycin and cefazedone, so both were used. E. coli
had resistance to only lincomycin, so only lincomycin resistant plasmid DNA was used. E.
coli's clear zone decreased from 3.4 cm of control to between 2.9 with transformation. S.
gallinarum control had 3.3 cm clear zone, but lincomycin plasmid DNA had 3 cm and
cefazedone 3.2 cm. The conclusion can be drawn that transformation by antibiotics resistant
plasmid DNA helps bacteria to acquire resistance, which is shown by decrease in clear zone.
Figure 7: Diameters of control and ABR transformed bacteria in co-cultivation
4. DISCUSSION
ABR spread through plasmid DNA; Experiment 5 clearly showed that implanting
ABR plasmid DNA can grant the bacteria with ABR trait. This ties closely with
transformation of bacteria. The question rises, however. What is the incentive for the bacteria
APEC Youth Scientist Journal Vol.7 / No.2
to accept the new DNA.
Strengthened ABR due to difference in types or concentrations of antibiotics;
Experiment 3 and 4 expressed that ABR can be strengthened by exposure to low
concentration of antibiotics or different antibiotics. Low concentration could not be explained
in perspective of transformation. Although it is antibiotics, low concentration would not kill
as many bacteria as higher concentration. This leads to lesser dead bacteria, which leads to
more surviving bacteria. With higher concentration of antibiotics Survival of the Fittest
would be the only factor, but besides those with inborn traits, those without traits cannot
acquire traits in anyway even with transformation because dead bacteria would not have any
useful gene for building defense against the antibiotics.
Preexisting ABR on old antibiotics; Experiment 1 and 2 showed that common bacteria
in soil and stored bacteria both had ABR to old antibiotics. This relates back to Charles
Darwin's Survival of the Fittest theory. When bacteria are exposed to antibiotics, many are
decimated, but few mutated bacteria survive. These bacteria inherit traits that make
antibiotics less effective, and the population grows with individuals with that trait, resulting
in ABR for the majority. Stored bacteria would not have problem developing ABR because
their origins are ultimately humans, who used antibiotics. The question is how common
bacteria exposed to antibiotics before did. One hypothesis is that disposal of antibiotics.
Antibiotics have form of liquid, so they were disposed in sink. This sink system is linked to
everywhere, so bacteria in soil first contacted the antibiotics. They formed and grew ABR
population spread the ABR through one or more of the methods above.
5. CONCLUSION
Experiment 1 and 2 showed that many bacteria have grown ABR to old antibiotics
and are growing ABR to newer ones. Experiment 3 through 5 proposed possible
mechanisms for ABR spread. The final conclusion is that ABR has been first introduced to
bacteria by exposure to low concentration of antibiotics, and this ABR has been spread by
proximity with ABR bacteria and transformation through ABR bacteria's plasmid DNA.
This spread was done for long time, which affected common bacteria as well, while bacteria
had less time to spread ABR for newer antibiotics, thus showing less ABR than to older

APEC Youth Scientist Journal Vol.7 / No.2
6. REFERENCES
Bergman, J . (2011, May 2). Bacteria and Antibiotics: An Example of Evolution by
Gustafsson, I., Sjölund, M., Torell, E., Johannesson, M., Engstrand, L., Cars, O.
Andersson, D. (2003). Bacteria with increased mutation frequency and antibiotic
resistance are enriched in the commensal flora of patients with high antibiotic usage.
Journal of Antimicrobial Chemotherapy, 52, 645-650.
Timeline of Antibiotic Resistance. (2015, April 14). Switchyard Media. Retrieved
Harjvan, C., Ho, A., and Farooqi, S. (2015, April 14). The Emergence of Novel
Pathogens: The Greatest Risk of Hubris in Human Health. PWC. Retrieved from
Houndt, T and Ochman, H. (2000) Long-Term Shifts in Patterns of Antibiotic
Resistance in Enteric Bacteria. Appl Environ Microbiol, 66(12), 5406–5409.
Stephens, E. (April 14, 2015). Types of Antibiotics. Emedicine health. Retrieved
A Brief Overview of Classes of Antibiotics. (2015, April 14). Compound Interest.
Retrieved from http://www.compoundchem.com/2014/09/08/antibiotics/.
New Hypothesis: Why Bacteria are becoming increasingly more Resistant to
Antibiotics. (2015, April 14). Phys.org. Retrieved from http://phys.org/news/2013-
An H-NS-like Stealth Protein Aids Horizontal DNA Transmission in Bacteria.
Science, 315 (5809), 251-252.
[10] Beaber, J. W., Hochnut, B. and Waldor, M. K. (2004). SOS response promotes
horizontal dissemination of antibiotic resistance genes. Nature, 427, 72-74.
[11] L. CLD. I. (2014).
Selection of a Multidrug Resistance Plasmid by Sublenthal Levels of Antibiotics and
Heavy Metals. Mbio, 5 (5), 01918-14.
Source: http://www.amgs.or.kr/New/common/journal/vol7/vol7_2_no.10.PDF
By: Frehiwot Mulugeta Seyoum Tefera Table of Contents 1. Introduction As in most developing countries in Ethiopia agriculture is the dominant sector of the economy. It contributes the lion share of the Gross Domestic Product (GDP) and foreign currency earnings of the country from the sell of agricultural outputs abroad. According to
Wednesday, November 12, 2008 9:00 a.m. – 9:30 a.m. 9:30 a.m.—10:00 a.m. Mark Hoyert, Interim Dean of the College of Arts and Sciences l0:15 a.m. - 11:45 a.m. Session I- Retention and Assessment of Student Learning, LCC 105AB Moderator: Karl Nelson, Department of Psychology, IU Northwest Assessing the Impact of Geoscience Laboratories on Student Learning. Karl Nelson, Department of Psychology, IU Northwest, Kristin Huysken, Department of Geosciences, IU Northwest, and Zoran Kilibarda, Department of Geosciences, IU Northwest


