Caracal.biz
Ó 2013 International Association for Ecology and Health
Original Contribution
Tracking Pathogen Transmission at the Human–WildlifeInterface: Banded Mongoose and Escherichia coli
R. Pesapane,1 M. Ponder,2 and K. A. Alexander1,3
1Department of Fish and Wildlife Conservation, Virginia Tech, Virginia Polytechnic Institute and State University,Fish and Wildlife Conservation, 152 Cheatham Hall (0321), Blacksburg, VA 240612Department of Food Science and Technology, Virginia Tech, Blacksburg, VA3CARACAL: Centre for Conservation of African Resources Animals, Communities and Land Use, Kasane, Botswana
Abstract: A primary challenge to managing emerging infectious disease is identifying pathways that allow
pathogen transmission at the human–wildlife interface. Using Escherichia coli as a model organism, we eval-
uated fecal bacterial transmission between banded mongoose (Mungos mungo) and humans in northern
Botswana. Fecal samples were collected from banded mongoose living in protected areas (n = 87, 3 troops) and
surrounding villages (n = 92, 3 troops). Human fecal waste was collected from the same environment (n = 46).
Isolates were evaluated for susceptibility to 10 antibiotics. Resistant E. coli isolates from mongoose were
compared to human isolates using rep-PCR fingerprinting and MLST-PCR. Antimicrobial resistant isolates
were identified in 57 % of the mongoose fecal samples tested (range 31–78% among troops). At least one
individual mongoose fecal sample demonstrated resistance to each tested antibiotic, and multidrug resistance
was highest in the protected areas (40.9%). E. coli isolated from mongoose and human sources in this study
demonstrated an extremely high degree of genetic similarity on rep-PCR (AMOVA, FST = 0.0027, p = 0.18)with a similar pattern identified on MLST-PCR. Human waste may be an important source of microbial
exposure to wildlife. Evidence of high levels of antimicrobial resistance even within protected areas identifies an
emerging health threat and highlights the need for improved waste management in these systems.
Keywords: zoonotic, Escherichia coli, banded mongoose, human waste, emerging infectious disease, antibiotic
reservoirs (Jones et al. ). Zoonotic disease is also anincreasing threat to humanity globally, crossing cultural
As human populations grow and transform landscapes,
and socioeconomic lines with developed and developing
contact with wildlife concomitantly increases. Disease
nations equally at risk (Mayer With increased travel
emergence has been an important consequence of this
and global interconnectedness, emergence of zoonotic
escalation in interaction, with the majority of emerging
disease in any one place has the potential to spark a global
infectious diseases in humans now arising from wildlife
pandemic, as seen with sudden acute respiratory syndrome(Dye and Gay ) and swine flu (H1N1 influenza)(Neumann et al. ). Indeed, some of the most devas-
Correspondence to: K. A. Alexander, e-mail:
[email protected]
R. Pesapane et al.
tating and persistent human pathogens can be traced to
Recent studies demonstrate the utility of Escherichia
zoonotic origins, (i.e., human immunodeficiency virus/
coli as a model for examining transmission of fecal
acquired immunodeficiency syndrome (Hahn et al. )
microorganisms between humans and wildlife (Singh
and influenza (Schoub
et al. Chapman et al. ; Skurnik et al.
Pathogen transmission from humans to wildlife is also
Goldberg et al. , ; Rwego et al.
an important emerging threat, increasing wildlife conser-
Farnleitner et al. Szekely et al. ). E. coli is a
vation and management challenges (Deem et al. For
bacterium found in the gastrointestinal tract and feces of
example, emergence of human metapneumovirus morbidity
all mammals. While the majority of E. coli are important
and mortality in mountain gorillas (Gorilla beringei berin-
commensals, some strains can be pathogenic (e.g.,
gei) in Rwanda is associated with human–gorilla contact
Enterotoxigenic E. coli) in humans, domestic animals, and
related to ecotourism activities (Palacios ). Similar
wildlife. For example, E. coli is one of the leading causes
studies demonstrate the increasing threat of human infec-
of diarrheal-related deaths worldwide (Gaedirelwe
tious diseases to non-human primate species (Nizeyi et al.
Drummond Stecher et al. ). While there are a
; Graczyk et al. ), elephants (Michalak et al.
number of limitations to the use of E. coli (Desmarais
sea birds (Steele et al. and reptiles (Wheeler et al.
et al. as a model organism, it is still regarded as
). These examples illustrate the bidirectional potential
the global standard for detection of fecal contamination,
for pathogen transmission at the human–wildlife interface.
indicating an increased possibility for the presence of
Human modification of the environment is seen as a
enteric pathogens.
primary driver of the emergence of zoonotic disease, pro-
In northern Botswana, humans live in close association
viding the opportunity for direct and indirect contact be-
with a variety of wildlife species. One species in particular is
tween humans and wildlife and increasing pathogen
the banded mongoose (Mungos mungo). Banded mongoose
exposure and transmission potential (Mayer ; Deem
occur across protected and unprotected landscapes with
et al. These changes can induce immediate as well as
significant contact with humans in this system, coupled
long-term effects on pathogen transmission dynamics,
with a preference for denning and foraging in anthropo-
modifying genetic and biological characteristics, biophysical
genic features (Alexander et al. ). Banded mongoose
elements, ecological dynamics, and socioeconomics, as well
also utilize garbage resources and will search for insects in
as host(s)–pathogen interactions (Smolinski et al.
fecal waste (including human) found in the environment.
Human fecal waste is an anthropogenic disturbance that
This species is also infected with two important emerging
persists across human-occupied landscapes, both in devel-
pathogens: the globally important zoonotic pathogen, lep-
oped and developing nations, serving as an important
tospirosis (Jobbins et al. ) and the newly discovered
source of environmental pathogens (Cilimburg et al. ;
Mycobacterium mungi, a member of the Mycobacterium
Griffin et al. ). Sewage sludge and refuse hosts an array
tuberculosis complex (Alexander et al. It is unclear at
of pathogenic bacteria, viruses, protozoa, and helminthes
present if M. mungi has the potential to infect humans or
that can lead to outbreaks of disease (Hanks ; Burge
other wildlife in the system. Banded mongoose's contact
and Marsh ; Arthurson ). Contamination of
with other wildlife and humans makes this species an ideal
drinking water by sewage effluent is a recurring cause of
candidate for evaluating microbial exchange at the human–
gastroenteritis leading to morbidity and mortality world-
wildlife interface.
wide (D'Antonio et al. ; Ashbolt This is espe-
We determined the level of antimicrobial resistance of
E. coli isolates collected from feces of banded mongoose
populations often lack regulated waste management and
occurring across protected and unprotected areas in
sanitation infrastructure. While environmental fecal waste
northern Botswana. We then evaluated the genetic rela-
may be an important source of pathogen exposure for both
tionships between antibiotic-resistant E. coli isolates from
wildlife and humans, we still have a limited understanding
mongoose and isolates from human fecal waste. We used
of the complex process of pathogen spillover between
these results to examine the potential for human fecal waste
wildlife and humans. The relative infrequency of pathogen
to act as a source gastrointestinal pathogen exposure for
spillover events limits our ability to evaluate the complexity
wildlife species living in both protected and unprotected
of interacting and cascading factors driving this process.

Tracking Pathogen Transmission
located within the Chobe National Park. In this region,there are no commercial chicken or livestock production
systems and agriculture is, therefore, expected to contributeminimally to the accumulation of antibiotic resistance in
This study was conducted in Chobe District in northern
the region. In this same area, we have established a long-
Botswana along the Chobe River, which traverses the Chobe
term ecological study of banded mongoose (Mungos mun-
National Park and the associated townships of Kasane and
go) in both protected and unprotected land areas (Alex-
Kazungula (Fig. ). Chobe District is 20, 800 km2 in size
ander et al. ).
with a relatively low population of *23,387 people (Cen-tral Statistics Office BG largely restricted to villages
surrounding the Chobe National Park. The rest of the landarea in the District is designated as protected (e.g., national
The banded mongoose host is a small diurnal viverrid,
parks, wildlife management areas, and forest reserves).
group-living, communal breeder, and a natural inhabitant
Chobe District has rich and diverse wildlife resources.
of wildlife areas in proximity to permanent water. They
Residential and tourism development occur near the pro-
occur in groups or troops that can number between 8 and
tected area network, as does a limited amount of subsis-
55 individuals and frequently cohabit with humans in
tence agriculture. A few permanent tourism facilities are
towns where they scavenge in human waste and feces.
Figure 1. Map of the study site. In northern Botswana, mongoose troops were sampled (triangles) in Chobe District in protected areas (Chobe
National Park and Kasane Forest Reserve, hatched outline, troops living in the protected area are denoted with ‘‘+'') and towns of Kasane and
Kazungula. The Chobe River flows through the Park to industrial, commercial, residential developments (gray circles) and commercial
agricultural areas (light gray polygons). The sewage treatment facility is located in the Kazungula area (stripes). Tourism facilities are found
within the developed and protected areas (stars). Sources of human waste such as feces and sewage (squares) found in the environment near
study troops were sampled as well as the sewage treatment facility.
R. Pesapane et al.
Banded mongoose have a low-skew reproductive strategy;
multiple females breed and there is little or no observed
suppression of breeding in subordinates (Gilchrist ).
Dispersal can be infrequent and individuals may be re-
tained within their natal group and breed with relatives
upon sexual maturity (Cant et al.
In our long-term study site, at least one or two animals
in each study troop are radio-collared and monitored
regularly with spatially referenced habitat, behavioral,
clinical, and other relevant ecological data collected on the
study population (Laver et al. Home range data were
used to identify mongoose troops as occurring in protected
areas or in surrounding villages. This study was conducted
under a permit from the Botswana Ministry of Environ-
ment, Wildlife, and Tourism and with approval of the
Virginia Tech's Institutional Animal Care and Use Com-
mittee (Protocol number 07-146-FIW and 10-154-FIW).
Fecal and Environmental Sample Collection
Fecal samples were collected from banded mongoose living
in protected areas (n = 87, 3 troops named CCL+, CGL+,
HP+ with ‘‘+'' denoting protected area) and surrounding
villages (n = 92, 3 troops, KUB, CSL, SEF, Fig. ). With the
exception of eight individuals in one troop in the protected
area (HP+), all troops had some level of range overlap with
human populations. The CCL+ and CGL+ troop homerange included tourism facilities within the protected areas
where some contact with humans and development would
be experienced but at a much lower level than in the village
troops. The HP+ troop occurred in a part of the protected
area where there was no human infrastructural development
or residences other than a dirt road. Feces from these 6
banded mongoose troops were collected following morning
latrine behavior. Banded mongoose defecate individually at
the same time period and in the same general location each
morning upon leaving their den; it is, therefore, possible to
collect fecal samples from individual mongoose in eachtroop without replication during that latrine event. In order
to determine the required interval between troop resam-
pling, such that each fecal sample could be considered an
independent sampling point for the troop, we measured gut
passage time. Colored dye was added to the diet of four
captive mongoose (owned by CARACAL Biodiversity
Center) and minimum gut fecal passage time was deter-
mined to be approximately three days. Based on these re-
sults, three sampling efforts were attempted at intervals of
not less than 2 weeks to ensure independence of sampling
Tracking Pathogen Transmission
events. In this study, we pooled across sampling events,considering each fecal sample an independent sample given
established clearance time and latrine behavior. Fecal sam-
ples were collected in sterile tubes shortly after defecation
and frozen at -20°C within 8 h of collection until pro-
cessing (Roesch et al. Mongoose fecal samples aresmall and friable with large amounts of insect material,making sectioning of mongoose feces difficult. Mongoose
also consume soil while foraging and thus sampling con-straints were relaxed and the whole fecal sample was taken
and used without regard to exterior or interior components.
Human feces were collected within mongoose home
ranges from bush latrines (e.g., on ground defecation) andsewage sludge or wastewater leakage identified in the study
area (Fig. ). Only sections of human feces that did notcome in contact with soil were sampled to avoid environ-
mental contamination. Additional samples of sewage were
collected from the local wastewater treatment facility. In thecase of liquid samples such as wastewater, sterile gauze was
swirled in the source to catch particulates along with a
water sample in the same sterile tube. Soil samples werecollected from areas undisturbed by apparent human or
animal waste and were used to provide a limited assessment
of natural levels of antimicrobial resistance in the envi-
Fecal and wastewater samples were homogenized in buf-
fered peptone (0.1% BPW Sigma-Aldrich, St. Louis, MO
USA), serially diluted 10-fold, and spread plated ontoMacConkey Agar (Becton Dickinson Company, FranklinLakes, NJ, USA) plates. After 24 h at 37°C, six putative
E. coli colonies were picked from each mongoose fecalsample. Sewage waste represented an array of individual
sources; therefore, 15 colonies were selected from human
fecal samples to be used in subsequent analyses. All putative
E. coli colonies were grown in tryptic soy broth (BectonDickinson Company) for further DNA extraction and
antibiotic susceptibility testing.
DNA was extracted from putative E. coli colonies using
standard methods. Genera-specific primers for malB were
used to confirm isolates as E. coli as previously described(Rekha et al. ). E. coli isolates were then tested for
susceptibility to 10 antibiotics: ampicillin (10 lg), chlor-
amphenicol (30 lg), ciprofloxacin (5 lg), doxycycline
(30 lg), gentamycin (10 lg), neomycin (30 lg), strepto-mycin (10 lg), tetracycline (30 lg), sulfamethoxazole–

R. Pesapane et al.
Hierarchical Analyses of Molecular Variance (AMOVA) for E. coli Isolates Collected from Humans and Mongoose in the
Chobe Region of Botswana.
Variance component
Observed partition
Mongoose and Humansa
HP+ versus CSL, CGL+, SEF, KUB, CCL+
Isolates were analyzed from banded mongoose troops (n = 6) and human bush latrine sites as well as sewage effluent in the environment within the homeranges of study mongoose. Banding patterns of bacterial genotypes were converted to binary variables, identified and scored using the ‘‘bandmatch''procedure in the FPQuest, version 4.5 program (BioRad). The program Arlequin, version 3.1 (Excoffier et al. ) was used to perform AMOVA.
aHuman samples consisted of feces (n = 5, 41 isolates) and sewage (n = 14, 46 isolates).
Figure 2. Occurrence of resis-
tant E. coli isolates in fecal
samples collected from individ-
ual banded mongoose by troop
in Chobe District, Botswana.
Mongoose troops occurred in
both the protected areas (+) and
surrounding villages. All error
bars represent 95% confidence
trimethoprim (SXT) (25 lg), and ceftiofur (30 lg) (BBL,
method with breakpoints indicated by the Clinical Labo-
Becton Dickinson Company). Antimicrobials were chosen
ratory Standards Institute (CLSI) In this modifi-
based on their local availability and on previous studies on
cation, a McFarland standard was not used to standardize
microorganism transmission from ecotourism (Rolland
turbidity. Instead, quality results were ensured in several
et al. Skurnik et al. ; Wheeler
ways. First, E. coli ATCC 25922 culture, a quality assurance
et al. ). Ceftiofur was added to the panel as it is only
indicator strain recommended by CLSI, was treated iden-
used in livestock and poultry production and was not
tically to sample isolates and results remained consistently
available in the study area. Antibiotic susceptibility was
within required limits for each test batch. Second,
measured using a field-modified Kirby-Bauer disk diffusion
any isolates demonstrating intermediate resistance were
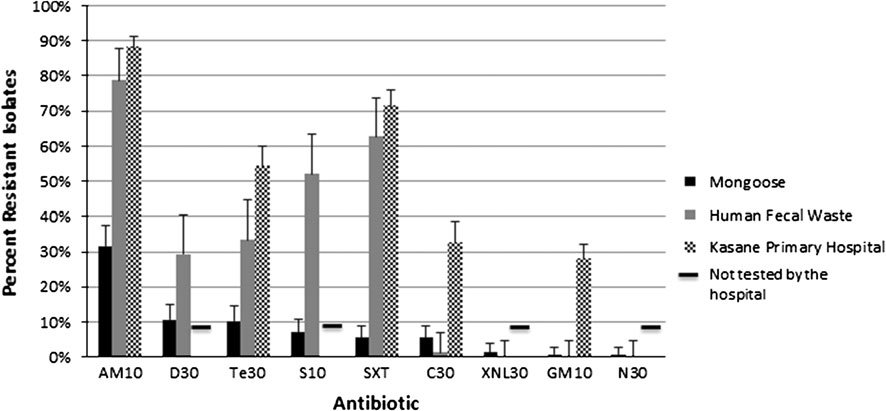
Tracking Pathogen Transmission
Figure 3. Prevalence of antibiotic resistance among total E. coli isolates collected from banded mongoose (n = 253), and human fecal waste
(n = 75) in Chobe District Botswana. Resistance data were also extracted from routine laboratory microorganism susceptibility
assessments conducted at the Kasane Primary Hosptial within the study site (2007–2011), on pre-established panels of antibiotics. Isolates
would originate from patients from all villages in the District. Not every isolate was subjected to the same panel of antibiotics at the hospital and
so the number of isolates screened for each drug varied. Antibiotics are listed as abbreviations followed by dosage (lg) in numbers, the number
of human isolates tested at the hospital are noted in brackets (e.g., AM10 = ampicillin [n = 420], Te30 = tetracycline [n = 315],
D30 = doxycycline, SXT = sulfamethoxazole-trimethaprim [n = 375], S10 = streptomycin, C30 = chloramphenicol [n = 256], XNL30 = cef-
tiofur, GM10 = gentamycin [n = 521], N30 = neomycin, and CIP5 = ciprofloxacin). Black bars denote antibiotics that were not evaluated at
health facilities in Chobe District during the assessment period.
accepted as susceptible for conservative reporting. Third,
lates were evaluated. Genetic relationships were determined
since stage of growth can affect resistance, all cultures tested
by repetitive polymerase chain reaction (rep-PCR) and
were >24 h of age (which corresponds to stationary phase)
multi-locus sequence-type polymerase chain reaction
in tryptic soy broth at the time of testing. We define
(MLST-PCR). In summary, rep-PCR reactions were per-
‘‘multidrug resistant'' isolates as those resistant to 3 or
formed in 25 lL reactions using 2 lL DNA template (of
more of the antibiotics screened for susceptibility.
standardized concentration 100–300 ng/lL) added to 2.5 U
Human antimicrobial resistance data for the region were
Phire Taq (Finnzymes) and 19 of 59 Phire Taq Buffer,
extracted from the local primary hospital (all data were
1 lM of BOX AIR primer (50-CTACGGCAAGGCGACG
anonymized, May 2007–July 2011). The research was con-
CTGACG-30) and an additional 1.5 mM of MgCl2. Cycling
ducted under permit from the Ministry of Health in Botswana
was performed using a BioRad MyCycler (Bio-Rad) at 98°C
and approval from the Virginia Tech Institutional Review
for 3 min, followed by 35 cycles of 98°C for 1 min, 64°C
Board (IRB# 11-573). Data were derived from routine testing
for 8 min, and 71°C for 1 min. Final extension was 15 min
of clinically isolated microorganisms from various fluids and
at 71°C. PCR fragments (2 lL DNA template of stan-
tested on pre-established antibiotic panels in order to deter-
dardized concentration 100–300 ng/lL) were separated by
mine susceptibility and treatment regimes. In this clinical
electrophoresis in a 2% agarose gel using 19 TAE buffer at
setting, intermediate susceptibility is reported as resistant to
45 V for 10 h at 4–8°C and visualized. Rep-PCR fingerprint
ensure the most appropriate drug treatment regime.
profiles were generated using densitometric (curve-based)genotype determination (Goldberg et al. with theFPQuest ver 4.5 program (BioRad, Hercules, CA). Genetic
Genetic Profiling of Mongoose and Human Bacte-
diversity of fecal E. coli isolates were assessed using analysis
rial E. coli Isolates
of molecular variance (AMOVA) in the Arlequin program
While all human E. coli isolates were analyzed for genetic
version 3.0 as previously described (Excoffier et al. ).
relatedness, only antibiotic-resistant mongoose E. coli iso-
Phylogenetic relationships were built through Pearson's
R. Pesapane et al.
correlation coefficient and neighbor-joining algorithms
Figure 4. Phylogram of rep-PCR fingerprint analysis of E. coli c
with optimization parameters identified from previously
isolates obtained from human and antibiotic-resistant banded
published guidelines (Goldberg et al. ).
mongoose fecal samples (n = 74 mongoose, n = 87 human).
To confirm the genetic relationships generated from
Comparisons were constructed using the Pearson's correlation and
neighbor-joining algorithms. Values alongside branches correspond
fingerprint analysis, a subset of isolates representing each
to % confidence generated from 1,000 bootstraps. Isolates that did
phylogenetic clade were amplified and sequenced for
not visibly differ were not included. Tree is rooted with the
MLST-PCR to construct a tree based on composite geno-
fingerprint profile of E. coli K12 from the EcMLST database.
types at different loci in a single isolate (Archie et al. Seven housekeeping loci (aspC, clpX, fadD, icdA, lysP, mdh,and uidA) were PCR amplified and sequenced as previously
production. Multidrug resistance (7.7–50%) was identified
described (STEC ). Resulting DNA sequences were
among a number of samples (Table Prevalence of mul-
aligned and phylogenetic analysis was performed using the
tidrug resistance among sampled mongoose feces was sig-
program Geneious 5.4.2 (Biomatters) (Drummond
nificantly different between sampled troops (p = 0.01, v2
Dendrograms were constructed using neighbor joining and
test). Troop KUB had the lowest level of multidrug resistance
pairwise similarity matrix methods. The tree was rooted
among individual fecal samples (7.7% with 95% CI 0.2–36%)
with composite sequence of the E. coli K12 from the
while troops SEF (50% with 95% CI 1.3–98.7% n = 2) and
EcMLST database ().
CGL+ (40.9% with 95% CI 20.7–63.6% n = 22) had thehighest levels. There were no significant differences in resis-
tance to two or fewer antimicrobials (p = 0.11) among fecalsamples from study troops. Troop CGL+, located within the
Fisher's exact and v2 tests were used to compare antibiotic
protected area, did not have the highest prevalence of resis-
resistance levels among groups using the statistical software
tant isolates overall (Fig. but was the only troop where at
JMP, version 8.0 (SAS Institute). Exact binomial 95%
least one fecal isolate was resistant to each of the antibiotics
confidence intervals were calculated using the Epitools
screened (Fig. Table Troop HP+ had the highest
package in R (Aragon and Enanoria
prevalence of antibiotic-resistant isolates although thesewere resistant to ampicillin only (Fig. Table ).
E. coli were isolated from 41.3% (n = 46, feces and
sewage) of human fecal waste samples assessed [human
E. coli Isolation and Antibiotic Susceptibility
feces found directly in the environment: (n = 5), sewagespill (n = 8), and sewage treatment pond samples (n = 6)].
E. coli were isolated from 49% (n = 179) of sampled mon-
From these samples, 77 E. coli isolates were screened for
goose feces. No E. coli isolates were obtained from soil
antibiotic susceptibility and 80.3% (95% CI 69.5–88.5%)
samples collected from environments expected to be free of
were found to be resistant to at least one antibiotic. Of
human or animal fecal contamination (n = 13). In mon-
human clinical samples screened at the local hospital,
goose fecal samples where E. coli were isolated, 57% dem-
89.9% (95% CI 87–92% for all years) of isolated micro-
onstrated antimicrobial resistance. Resistance was identified
organisms (various sources and bacteria species) were
among individuals in all sampled troops (31–78%, Table
resistant to at least one antibiotic. Human and banded
No significant differences were observed in the prevalence of
mongoose E. coli isolates collected in this study demon-
antimicrobial resistant E. coli isolates across fecal sampling
strated resistance to the same drugs as microorganisms
events for each respective troop (p > 0.05) and data were
isolated from patients at the Kasane Primary Hospital
pooled by troop for analysis. At least one or more mongoose
(Fig. Overall, resistance in banded mongoose was lower
fecal samples demonstrated resistance to each antibiotic
than that found in the human population.
tested in this study. Mongoose fecal isolates were mostcommonly resistant to ampicillin (AM10), followed by
Repetitive BOX-PCR
doxycyline (D30), tetracycline (Te30), streptomycin (S10),SXT, and chloramphenicol (C30) (Fig. Table ). A low
Using AMOVA, significant variation was identified among
number of isolates (n = 5) demonstrated resistance to cef-
sampled mongoose feces (FST = 0.02, p = 0.01, Table ),
tiofur (XNL30), an antibiotic used in livestock and poultry
with the majority of the variation identified at the indi-

Tracking Pathogen Transmission
vidual fecal sample level (98% of observed variation) rather
living in an unmodified habitat (HP+) were indistin-
than at the troop level (2% of the observed variation).
guishable genetically from those troops living in association
However, E. coli strains isolated from the mongoose troop
with humans in villages or at lodges in protected areas

R. Pesapane et al.
Figure 5. Phylogenetic tree based
on E. coli composite sequences from
seven housekeeping genes (uidA,
mdh, lysP, icdA, clpX, aspC, and
fadD) (n = 23). Values alongside
branches correspond to % confi-
dence generated from 1,000 boot-
were built from seven consensus
trees using Tamura-Nei distance
model and neighbor-joining algo-
rithms, and 70% support threshold.
The tree is rooted with composite
sequence of E. coli K12 from the
Scale is based on the number
of base substitutions per site.
(CSL, CGL+, SEF, KUB, CCL+, FST = 0, p = 0.64,
rep-PCR analysis revealed very slight genetic divergence
Table When comparing humans and mongoose isolates,
(maximum < 0.10 base substitutions per site) between
a greater level of genetic diversity of E. coli was found
isolates from banded mongoose and human fecal waste
within each species (99.75% of the observed variation) than
(Fig. E. coli isolated from banded mongoose and sources
among species (0.25% of the observed variation). Mon-
of human fecal waste consistently clustered together,
goose and human associated E. coli demonstrated an
forming one monophyletic tree. Genetic relationships
extremely high degree of genetic similarity (FST = 0.0027,
between isolates interpreted through branching patterns
were not identical between fingerprint- and DNA sequence-
Antibiotic-resistant isolates from all troops of mon-
based analyses in this study, even when using optimization
goose consistently clustered with those of humans and
parameters published by previous studies (Goldberg et al.
human fecal waste in rep-PCR analysis (Fig. ). Clades
containing exclusively mongoose or human isolates werenot observed. Mongoose isolates did not cluster signifi-cantly by troop, but instead were highly intermixed with
isolates from human sources.
Using molecular strain comparisons, E. coli isolated fromfeces of banded mongoose troops demonstrated an
extremely high degree of genetic similarity to isolates
MLST-PCR performed on a subset of E. coli isolates
obtained from human fecal waste in these environments.
(n = 23) representing each of the clades observed in
This was identified among troops with home ranges in both
Tracking Pathogen Transmission
protected and unprotected land areas. Other studies have
greater diversity of drug resistance was identified and a
applied similar molecular techniques demonstrating the
higher prevalence of multidrug resistance occurred (Fig.
potential for microorganism exchange to occur between
Table ). Ampicillin resistance can be naturally acquired
humans, wildlife, and livestock sharing the same land area.
from environmental sources (Martinez ). Patient iso-
Comparisons between populations focused on the direct
lates from the local hospital were also frequently resistant to
collection of fecal material from human volunteers living in
ampicillin (Fig. ). Other studies have observed a gradient
these environments (Goldberg et al. ;
effect of antimicrobial resistance among environments and
). While these examinations have provided substantial
animals in relation to human proximity (Skurnik et al.
insight into the occurrence of microbial exchange at the
Allen et al. ; Wheeler et al. ). Our study
human–animal interface, the specific pathways where gas-
design did not allow for this type of evaluation; however,
trointestinal microbial transmission might occur remain
our limited data suggest that this might be an important
elusive. In this study, we focused our sampling strategy on
area of future work.
environmental sources of human fecal waste and compared
Multidrug resistance is increasingly identified as an
isolated E. coli to that of isolates collected from mongoose
important global health threat, involving all major micro-
living in these same areas.
bial pathogens and antimicrobial drugs. Multidrug resis-
Recent studies identify a high degree of genetic simi-
tance was identified among isolates collected from troops
larity among E. coli strains globally, regardless of human or
living in proximity to humans, both in the National Park
animal origin (Clermont et al. and suggest that
and in village areas. Of these, CGL+ (Park troop) had the
microbial exchange events implied by molecular genetic
highest level, with half of all sampled mongoose demon-
techniques should be viewed conservatively. However, in
strating multidrug resistance among isolates. In some
this study, we did find significant variation in E. coli by
troops, multidrug-resistant isolates were aggregated to a
species (human and mongoose, respectively). Our results
few individual mongoose fecal samples (e.g., KUB,
provide direct support for the possibility that direct human
Kazungula Village), but in the CGL+ troop, multidrug
fecal contamination of the environment is an important
resistance was widespread across individuals and isolates
potential source of microbial exposure and transmission to
(40.9%, 95% CI 20.7–63.6%). This troop commonly den-
wildlife living in these areas. This transmission appears to
ned in the soak away of the septic tank servicing the
occur even in protected wildlife areas.
employee accommodations and foraged around employee
Antimicrobial resistance is an important marker for
living quarters, including eating food remains from dishes
human-source bacteria (Kabler et al. ) and previous
left outside. Humans have also been observed feeding raw
studies identified antimicrobial resistance in wild popula-
meat waste from commercially produced chickens to
tions living in association with human populations (Cole
mongoose at this lodge, potentially contributing to the
et al. Skurnik et al. ; Goldberg et al. ;
levels of drug resistance in this troop. Antibiotic resistance
Wheeler et al. ). In this study, we found
to ampicillin and tetracycline has been identified in strains
high levels of antimicrobial resistance including multidrug
of Salmonella at commercial chicken farms in Botswana
resistance among sampled mongoose across all sample
(Gaedirelwe ) and may be an important source of
troops irrespective of land use (protected and unprotected).
resistant microbiota to species fed raw animal byproducts.
Important in evaluating the significance of the anti-
Alternatively, this resistance could be present in the human
microbial resistance identified in wildlife is evaluating the
population because of consumption of meat with antibiotic
occurrence in the resident human population. In this study,
residues. Wildlife could then be subsequently exposed
resistance to the same antimicrobials were identified among
through human fecal waste. Either or both of these
mongoose isolates (although lower levels) as that identified
mechanisms may have contributed to the low occurrence of
in study-derived human E. coli isolates and patients pre-
resistance to ceftiofur (XNL30, Table a broad-spectrum
senting to the Kasane Primary Hospital (2007–2011,
veterinary antibiotic not available in the study area. All of
Fig. Most isolates obtained from the mongoose troop
the study troops, with the exception of HP + , utilized
living in a natural area of Chobe National Park (HP+) were
garbage resources and would have access to such waste
resistant to only ampicillin, with one isolate demonstrating
albeit at varying levels depending on the facility. Uncon-
additional drug resistance. This result was in contrast to the
trolled and uncontained waste disposal provides a ubiqui-
other sample troops living in contact with humans where a
tous non-seasonal food resource in both peri-urban
R. Pesapane et al.
environments and protected areas, the latter in association
bial exchange and antibiotic resistance accumulation in
with ecotourism enterprises (Rolland et al. While
mongoose may extend through food webs. Mongoose are
ecotourism developments are important for conservation
eaten by a large number of avian, reptile, and mammalian
and economic growth, associated human waste (both gar-
predators including domestic dogs in this system. Thus, the
bage and feces) remains a primary negative consequence. It
cascading effects of exposure of wildlife species to human
is unclear how garbage utilization may contribute to
waste-associated microbes can impact an array of suscep-
microbial exchange and accumulation of antimicrobial
tible species across an ecosystem and in turn increase
resistance. Given the increasing nature of this anthropo-
human exposure, coupling humans and natural systems in
genic landscape modification, it is a crucial area for con-
complicated ways.
tinued research.
Human fecal waste and garbage are widespread across
There was a high level of resistance among human
many environments and are an increasingly global threat to
isolates collected in this study and those tested at the pri-
animal and public health (Rushton Cantalupo et al.
mary hospital. This may appear surprising at first given
This phenomenon extends to protected areas, where
poverty and expectations of diminished access to health
increasing pressure to develop economic enterprises based
care and pharmaceuticals that is assumed for much of
on tourism brings people and their waste even to remote
Africa. However, antibiotics are widely available to patients
areas of a protected area network. In order to control the
visiting government hospitals in Botswana, where health
threat of potential pathogen transmission, closed sewage
care is largely free, as well as local pharmacies where few
systems should be required. Feeding of poultry and live-
controls on the dispensing of antibiotics are identified.
stock products from kitchen waste should be actively pro-
Decreased regulation of antibiotic use and access is an
hibited to decrease the potential for transmission of
important contributor to the epidemic spread of resistance
resistant bacteria. Waste receptacles need to be completely
(Okeke et al. , and has likely contributed to
enclosed and ‘‘wildlife-proofed'' to discourage wildlife
the occurrence of increased microbial resistance in this
access and exposure.
human population.
Transmission of pathogens through fecal waste is likely
A number of study limitations could have influenced
to be amplified in places such as Africa, where sanitation
our results. For example, the potential for contamination of
and water treatment facilities are often poor, and fecal
sewage samples with non-human fecal material and soil is a
contamination of the environment and limited surface
possibility. It is likely, however, that such contamination, if
water resources represents a growing problem. Common
it occurred, would be present at exceptionally low levels
water resources may represent an important exposure
relative to the volume of human sewage. Sewage and direct
mechanism, increasing the potential for bidirectional
fecal contamination of the only surface water in the region
microbial traffic and increased health risks to both humans
creates another vehicle for microorganism transmission
and wildlife living in these systems. The intimate connec-
between humans and wildlife and could be an important
tions between human and animal populations are
mechanism of E. coli exchange between humans and
increasingly evident, as is the need to better understand
mongoose. Antibiotics from local human use may accu-
these system coupling points where human and animal
mulate in both the soil (Tolls ) and contaminated
health are interlinked.
surface water (Kummerer potentially increasing thespread of antibiotic resistance.
We would like to thank the Botswana Ministry of Health andthe Ministry of Environment Wildlife, and Tourism for
Our study suggests that environmentally associated human
permission to conduct this study. We thank M. Vandewalle
fecal waste may serve as an important source of microbial
and P. Laver for their invaluable assistance in this study and
transmission to mongoose. Furthermore, widespread
E. Hallerman and J. Fox for comments and assistance on
accumulation of antimicrobial resistance is identified
this manuscript. This project was funded under the
among mongoose even in protected areas considered to be
National Science Foundation Coupled Human Environmental
relatively free from human impacts. The impact of micro-
Systems Award CNH#1114953, Morris Animal Foundation
Tracking Pathogen Transmission
Award # D10Z0-828A, and the WildiZe Foundation. The
Desmarais TR, Solo-Gabriele HM, et al. (2002) Influence of soil
National Science Foundation S-STEM Program (DUE-0850198)
on fecal indicator organisms in a tidally influenced subtropicalenvironment.
provided partial financial support for R. Pesapane.
Drummond AJ, A. B., Buxton S, Cheung M, Cooper A, Duran C,
Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas
S, Sturrock S, Thierer T, Wilson A (2010). Geneious ver. 5.1.
Dye C, Gay N (2003) Epidemiology. Modeling the SARS epi-
demic. Science 300(5627):1884–1885
Alexander KA, Laver PN, et al. (2010) Novel Mycobacterium
Excoffier L, Smouse P, et al. (1992) Analysis of molecular variance
tuberculosis complex pathogen, M. mungi. Emerging infectious
inferred from metric distances among DNA haplotypes: appli-
diseases 16(8):1296
cation to human mitochondrial DNA restriction data. Genetics
Allen HK, Donato J, et al. (2010) Call of the wild: antibiotic
resistance genes in natural environments. Nature Reviews
Farnleitner AH, Ryzinska-Paier G, et al. (2010) Escherichia coli and
enterococci are sensitive and reliable indicators for human,
Aragon TJ, Enanoria WT (2007) Applied Epidemiology Using R,
livestock and wildlife faecal pollution in alpine mountainous
MedEpi Publishing.
water resources. Journal of Applied Microbiology 109(5):1599–
Calendar Time. Accessed 10 June 2010
Archie EA, Luikart G, et al. (2009) Infecting epidemiology with
Gaedirelwe OGaTKS (2008) The prevalence and antibiotic sus-
genetics: a new frontier in disease ecology. Trends in Ecology and
ceptibility of Salmonella sp in poultry and ostrich samples from
Evolution 24(1):21–30
slaughter houses in Gaborone, Botswana. Journal of Animal and
Arthurson V (2008) Proper sanitization of sewage sludge: a critical
Veterinary Advances 7(9):1151–1154
issue for a sustainable society. Applied Environmental Microbi-
Gilchrist JS (2006) Reproductive success in a low skew, communal
ology 74(17):5267–5275
breeding mammal: the banded mongoose, Mungos mungo.
Ashbolt NJ (2004) Microbial contamination of drinking water and
Behavioral Ecology and Sociobiology 60(6):854–863
disease outcomes in developing regions. Toxicology 198(1):229–
Goldberg TL, Gillespie TR, et al. (2008) Forest fragmentation and
bacterial transmission among nonhuman primates, humans,
Burge W, Marsh P (1978) Infectious disease hazards of land-
and livestock, Uganda. Emerging Infectious Diseases 14(9):1375–
spreading sewage wastes. Journal of Environmental Quality
Goldberg TL, Gillespie TR, et al. (2007) Patterns of gastrointes-
Cant M, Otali E, et al. (2002) Fighting and mating between groups
tinal bacterial exchange between chimpanzees and humans
in a cooperatively breeding mammal, the banded mongoose.
involved in research and tourism in western Uganda. Biological
Cantalupo PG, Calgua B, et al. (2011) Raw sewage harbors diverse
Goldberg TL, Gillespie TR, et al. (2006) Optimization of analytical
viral populations. mBio 2(5):1–11
parameters for inferring relationships among Escherichia coliisolates from repetitive-element PCR by maximizing corre-
Central Statistics Office BG (2011) 2011 Botswana Population and
spondence with multilocus sequence typing data. Applied and
Housing Census, Alphabetical Index of Districts.
Environmental Microbiology 72(9):6049–6052
. Accessed 31 March 2013
Graczyk TK, Bosco-Nizeyi J, et al. (2002) A single genotype of
Encephalitozoon intestinalis infects free-ranging gorillas and
Chapman CA, Speirs ML, et al. (2006) Life on the edge: gastro-
people sharing their habitats in Uganda. Parasitology Research
intestinal parasites from the forest edge and interior primate
groups. American Journal of Primatology 68(4):397–409
Griffin DW, Donaldson KA, et al. (2003) Pathogenic human
Cilimburg A, Monz C, et al. (2000) Wildland recreation and
viruses in coastal waters. Clinical Microbiology Reviews 16(1):
human waste: a review of problems, practices, and concerns.
Environmental Management 25(6):587–598
Hahn BH, Shaw GM, et al. (2000) AIDS as a zoonosis: scientific
Clermont O, Olier M, et al. (2011) Animal and human pathogenic
and public health implications. Science 287(5453):607–614
Escherichia coli strains share common genetic backgrounds.
Infection, Genetics and Evolution 11(3):654–662
Hanks T (1967) Solid waste/disease relationships, A literature sur-
vey, Cincinnati: U.S. National Center for Urban and Industrial
Clinical and Laboratory Standards Institute (2006) Performance
standards for antimicrobial disk susceptibility, Wayne, PA: Clin-ical Laboratory Standards Institute
Jobbins SE, Sanderson CE, et al. (2013) Leptospirosis in banded
mongoose at the human—wildlife interface in northern Bots-
Cole D, Drum DJ, et al. (2005) Free-living Canada geese and
wana. Zoonoses and Public Health (accepted)
antimicrobial resistance. Emerging Infectious Diseases 11(6):935–938
Jones K, Patel N, et al. (2008) Global trends in emerging infectious
diseases. Nature 451(7181):990–993
D'Antonio RG, Winn RE, et al. (1985) A waterborne outbreak of
cryptosporidiosis in normal hosts. Annals of Internal Medicine
Kabler PW, Clark HF, et al. (1964) Sanitary significance of coli-
form and fecal coliform organisms in surface water. PublicHealth Reports 79:58–60
Deem SL, Karesh WB, et al. (2001) Putting theory into practice:
wildlife health in conservation. Conservation Biology 15(5):
Kummerer K (2009) Antibiotics in the aquatic environment—a
review—part I. Chemosphere 75(4):417–434
R. Pesapane et al.
Laver P, Ganswindt A, et al. (2012) Non-invasive monitoring of
Rushton L (2003) Health hazards and waste management. British
glucocorticoid metabolites in banded mongooses (Mungos
Medical Bulletin 68:183–197
mungo) in response to physiological and biological challenges.
Rwego IB, Gillespie TR, et al. (2008) High rates of Escherichia coli
General and Comparative Endocrinology 179:178–183
transmission between livestock and humans in rural Uganda.
Martinez JL (2009) The role of natural environments in the
Journal of Clinical Microbiology 46(10):3187–3191
evolution of resistance traits in pathogenic bacteria. Proceedings
Rwego IB, Isabirye-Basuta G, et al. (2008) Gastrointestinal bac-
of the Royal Society: Biological Sciences 276(1667):2521–2530
terial transmission among humans, mountain gorillas, and
Mayer J (2000) Geography, ecology and emerging infectious dis-
livestock in Bwindi Impenetrable National Park, Uganda.
eases. Social Science and Medicine 50(7–8):937–952
Conservation Biology 22(6):1600–1607
Michalak K, Austin C, et al. (1998) Mycobacterium tuberculosis
Schoub BD (2012) Zoonotic diseases and human health: the
infection as a zoonotic disease: transmission between humans
influenza example. Onderstepoort Journal of Veterinary Research
and elephants. Emerging Infectious Diseases 4(2):283–287
Neumann G, Noda T, et al. (2009) Emergence and pandemic
Singh DK, McCormick C, et al. (2002) Pathogenic and nef-
potential of swine-origin H1N1 influenza virus. Nature
interrupted simian-human immunodeficiency viruses traffic to
the macaque CNS and cause astrocytosis early after inoculation.
Nizeyi JB, Innocent RB, et al. (2001) Campylobacteriosis, salmo-
Virology 296(1):39–51
nellosis, and shigellosis in free-ranging human-habituated
Skurnik D, Ruimy R, et al. (2006) Effect of human vicinity on
mountain gorillas of Uganda. Journal of Wildlife Diseases 37(2):
antimicrobial resistance and integrons in animal faecal Esche-
richia coli. Journal of Antimicrobial Chemotherapy 57(6):1215–
Okeke IN, Klugman KP, et al. (2005) Antimicrobial resistance in
developing countries. Part II: strategies for containment. The
Smolinski M, Hamburg M, et al. (2003) Microbial threats to health:
Lancet Infectious Diseases 5(9):568–580
emergence, detection, and response, Washington D.C.: The
Okeke IN, Lamikanra A, et al. (1999) Socioeconomic and
National Academies Press
behavioral factors leading to acquired bacterial resistance to
STEC (2011) Accessed 1 June 2010
antibiotics in developing countries. Emerging Infectious Diseases
Stecher B, Denzler R, et al. (2012) Gut inflammation can boost
horizontal gene transfer between pathogenic and commensal
Okeke IN, Laxminarayan R, et al. (2005) Antimicrobial resistance
Enterobacteriaceae. Proceedings of the National Academy of Sci-
in developing countries. Part I: recent trends and current status.
ences of the United States of America 109(4):1269–1274
The Lancet Infectious Diseases 5(8):481–493
Steele CM, Brown RN, et al. (2005) Prevalences of zoonotic
Palacios G (2011) Human metapneumovirus infection in wild
bacteria among seabirds in rehabilitation centers along the
mountain gorillas, Rwanda. Emerging Infectious Diseases 17(4):
Pacific Coast of California and Washington, USA. Journal of
Wildlife Diseases 41(4):735–744
Rekha R, Alam Rizvi M, et al. (2006) Designing and validation of
Szekely BA, Singh J, et al. (2010) Fecal bacterial diversity of hu-
genus-specific primers for human gut flora study. Electronic
man-habituated wild chimpanzees (Pan troglodytes schweinfur-
Journal of Biotechnology 9:505–511
thii) at Mahale Mountains National Park, Western Tanzania.
Roesch LF, Casella G, et al. (2009) Influence of fecal sample
American Journal of Primatology 72(7):566–574
storage on bacterial community diversity. Open Microbiology
Tolls J (2001) Sorption of veterinary pharmaceuticals in soils: a
Journal 3:40–46
review. Environmental Science and Technology 35(17):3397–3406
Rolland RM, Hausfater G, et al. (1985) Antibiotic-resistant bac-
Wheeler E, Hong PY, et al. (2012) Carriage of antibiotic-resistant
teria in wild primates—increased prevalence in baboons feeding
enteric bacteria varies among sites in Galapagos reptiles. Journal
on human refuse. Applied and Environmental Microbiology
of Wildlife Diseases 48(1):56–67
Source: http://caracal.biz/Our_publications_files/Pesapane,%20Ponder,%20and%20Alexander%202013%20Tracking%20Pathogens.pdf
LA CONSTITUCIÓN EUROPEA Y EL ESTATUTO JURÍDICO DE LAS REGIONES ULTRAPERIFÉRICAS MARÍA ASUNCIÓN ASÍN CABRERA Profesora titular de Derecho Internacional Privado Universidad de La Laguna Análisis del régimen jurídico de las regiones ultraperiféri- cas en el Tratado por el que se establece una Constitución A) La consolidación del modelo de integración de las
Peninsula Health - Community Dental Program Dental Management of Presentation based upon DHSV Clinical Practice Guidelines Presented by Dr Fayed Azouz Clinical Head Peninsula Health – Community Dental Program 1) To inform the Peninsula Health Community Dental clinical staff on the Current DHSV Clinical Practice Guidelines for dental care management of the pregnant woman.




